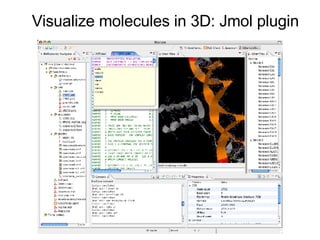
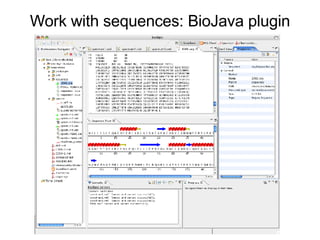
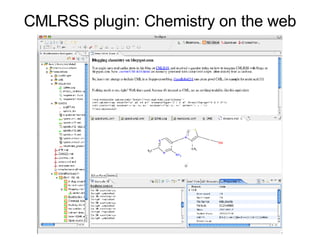
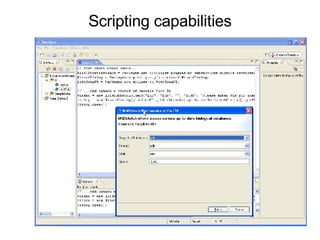
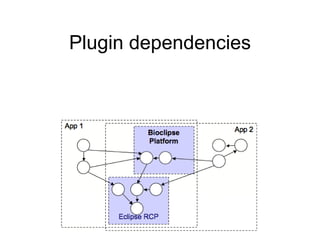
Bioclipse is an open source workbench for chemo- and bioinformatics that provides a platform for application development in life sciences. It integrates various functionality into a user-friendly workbench and is built on the Eclipse Rich Client Platform using a powerful plugin architecture. Bioclipse allows users to load, retrieve, edit, convert, analyze and visualize biological entities like small molecules, proteins and sequences.