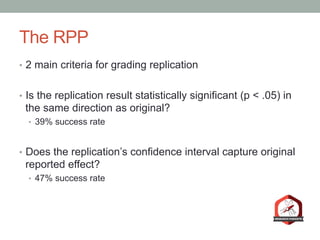
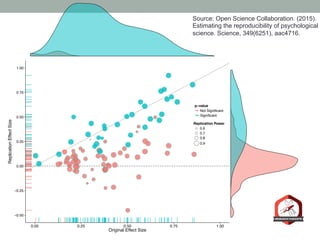
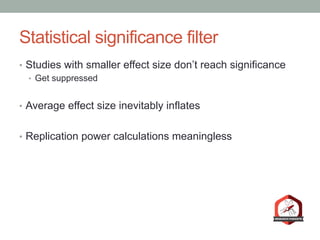
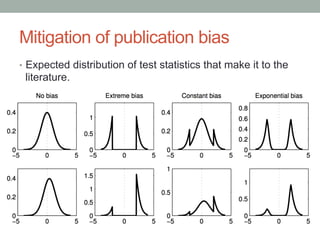
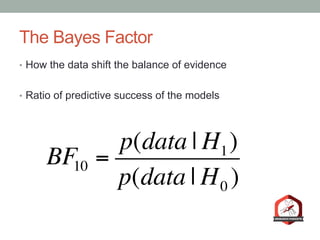
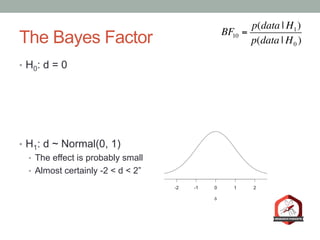
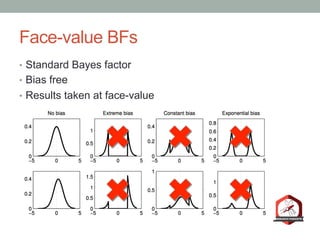
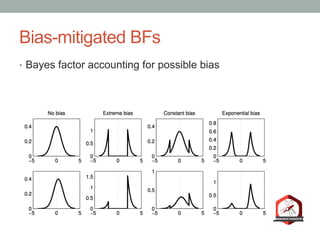
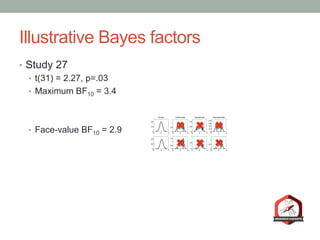
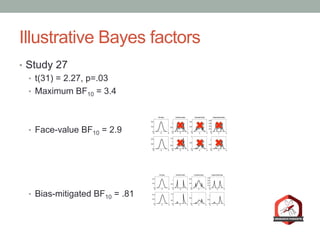
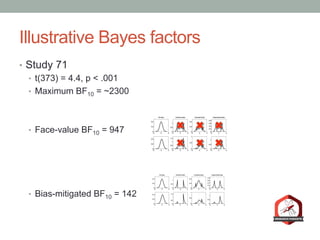
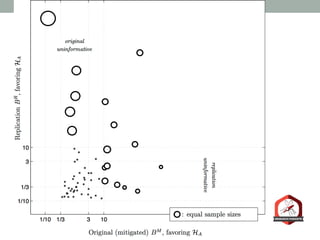
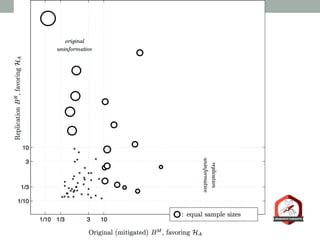
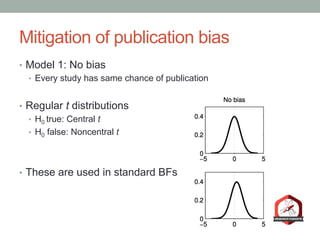
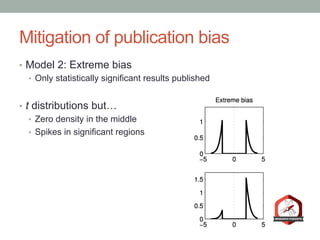
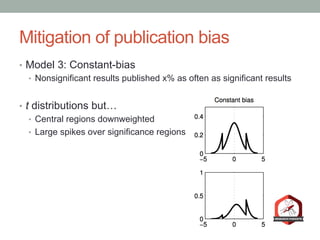
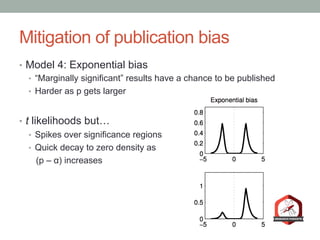
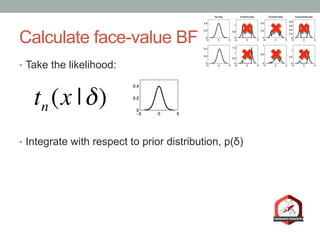
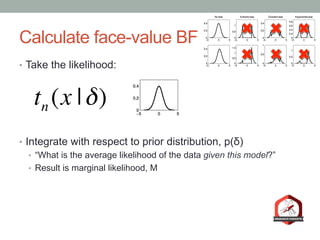
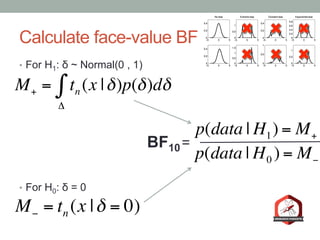
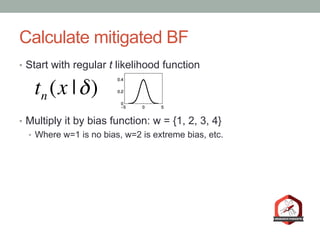
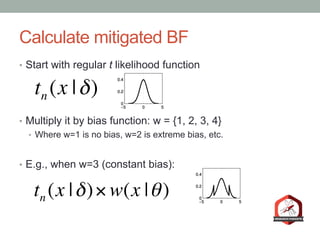
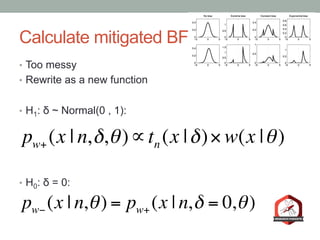
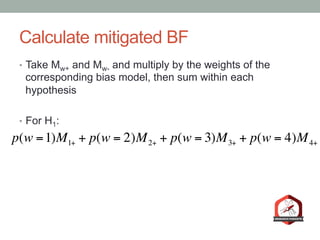
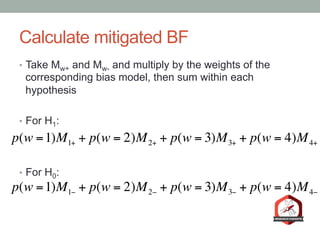
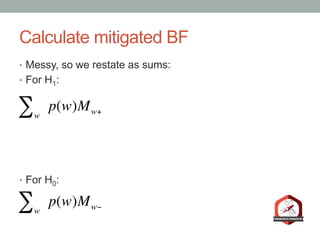
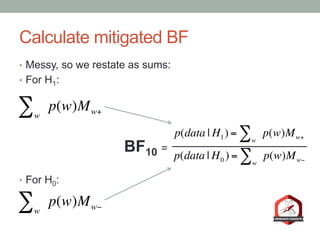
The document presents a Bayesian analysis of the Reproducibility Project: Psychology, which aimed to replicate original psychology studies and evaluate their statistical evidence. Findings indicate that 75% of studies show similar evidence in original and replication attempts, but the overall success rate of achieving significant results is relatively low. The analysis also addresses issues of publication bias and proposes Bayesian methods to account for this bias in interpreting evidence across studies.