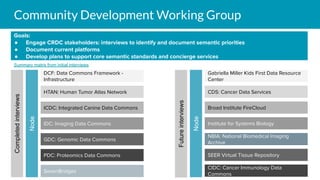
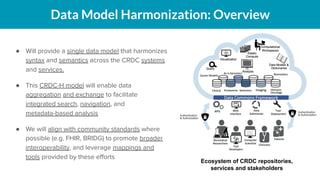
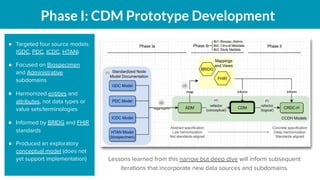
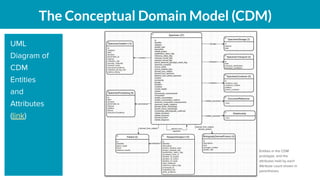
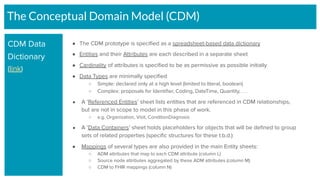
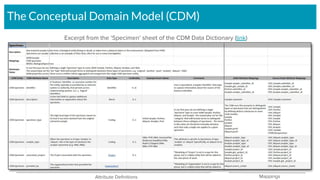
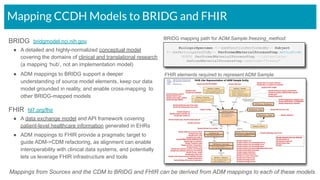
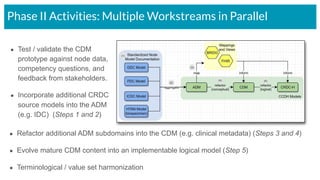
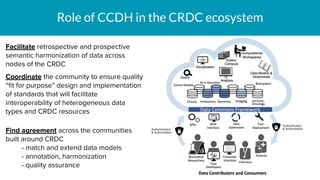
The document outlines the joint meeting of the Cancer Research Data Commons (CRDC) and the Center for Cancer Data Harmonization (CCDH) held on June 29, 2020, focusing on data model harmonization and community engagement strategies. It details workstreams aimed at developing a unified data model that ensures semantic interoperability across various cancer data platforms and provides insights into the methodologies for data preparation and quality assurance. Key outcomes include the development of an aggregated data model (ADM) and a conceptual domain model (CDM) designed to enhance data accessibility and usability for various stakeholders.




![Data Model
harmonization
(Lead: Chute
Co-Lead: Furner)
Ontology &
Terminology
Ecosystem
(Lead: Solbrig)
Tools & Data
Quality
(Lead: Balhoff)
Schema to
schema
OMOP to
FHIR
Term to
Term
Oncotree to
NCIt
Data records to
data records
“Smoking status 3
packs per day” to
NCIT:C154510
[Heavy Smoker]](https://image.slidesharecdn.com/crdcjunemeeting2020-200923164627/85/An-Introduction-to-CCDH-6-320.jpg)