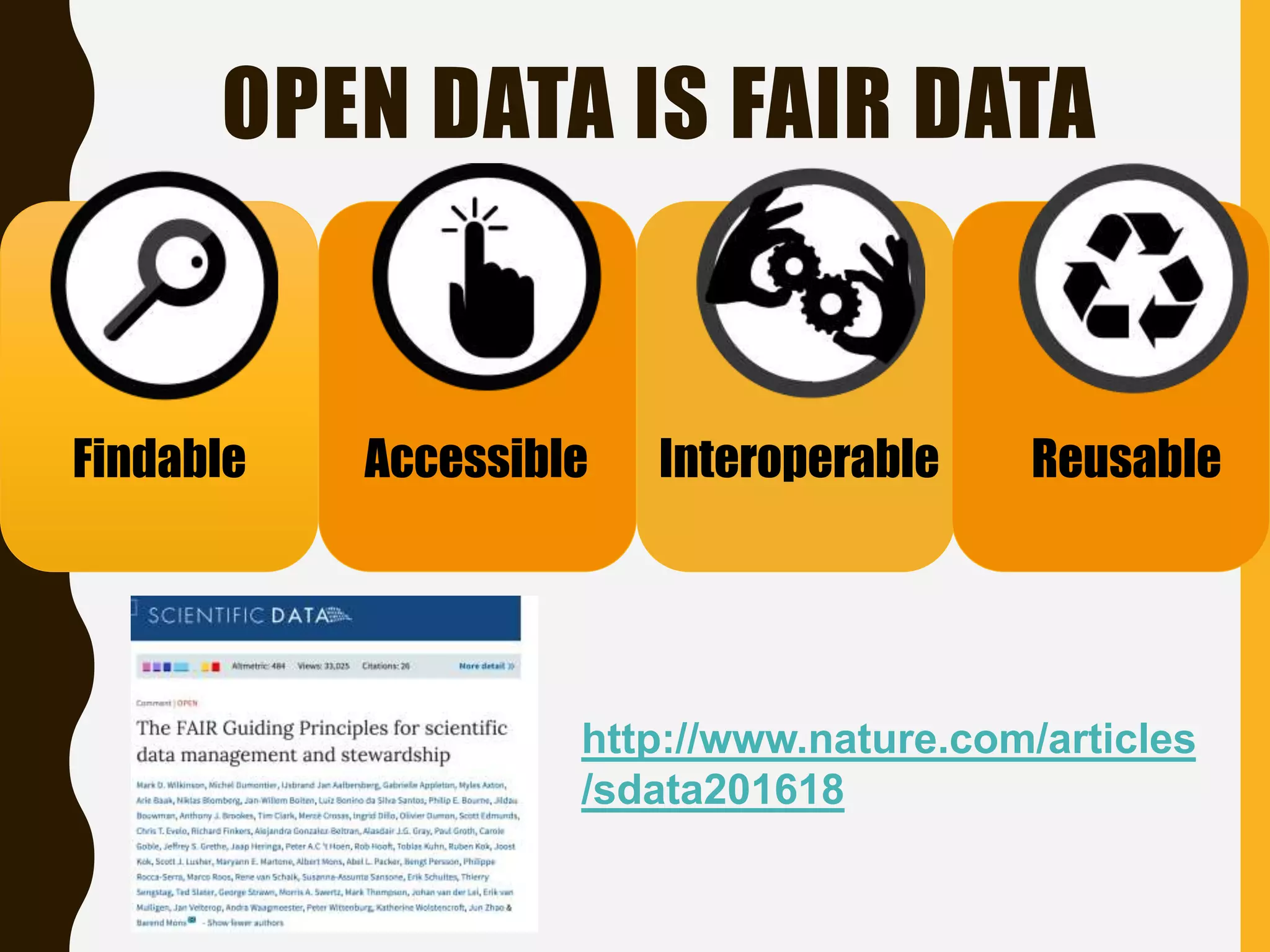
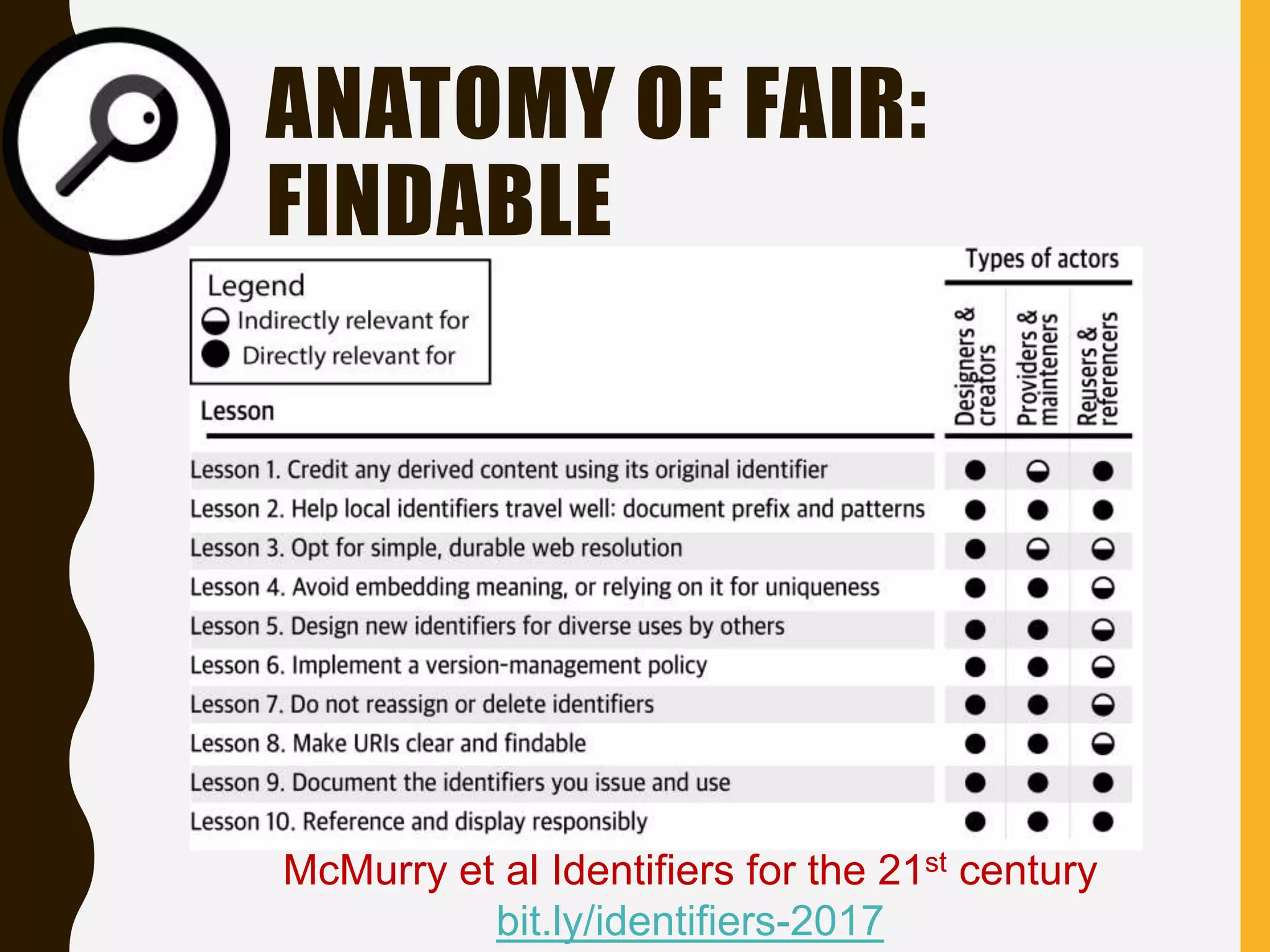
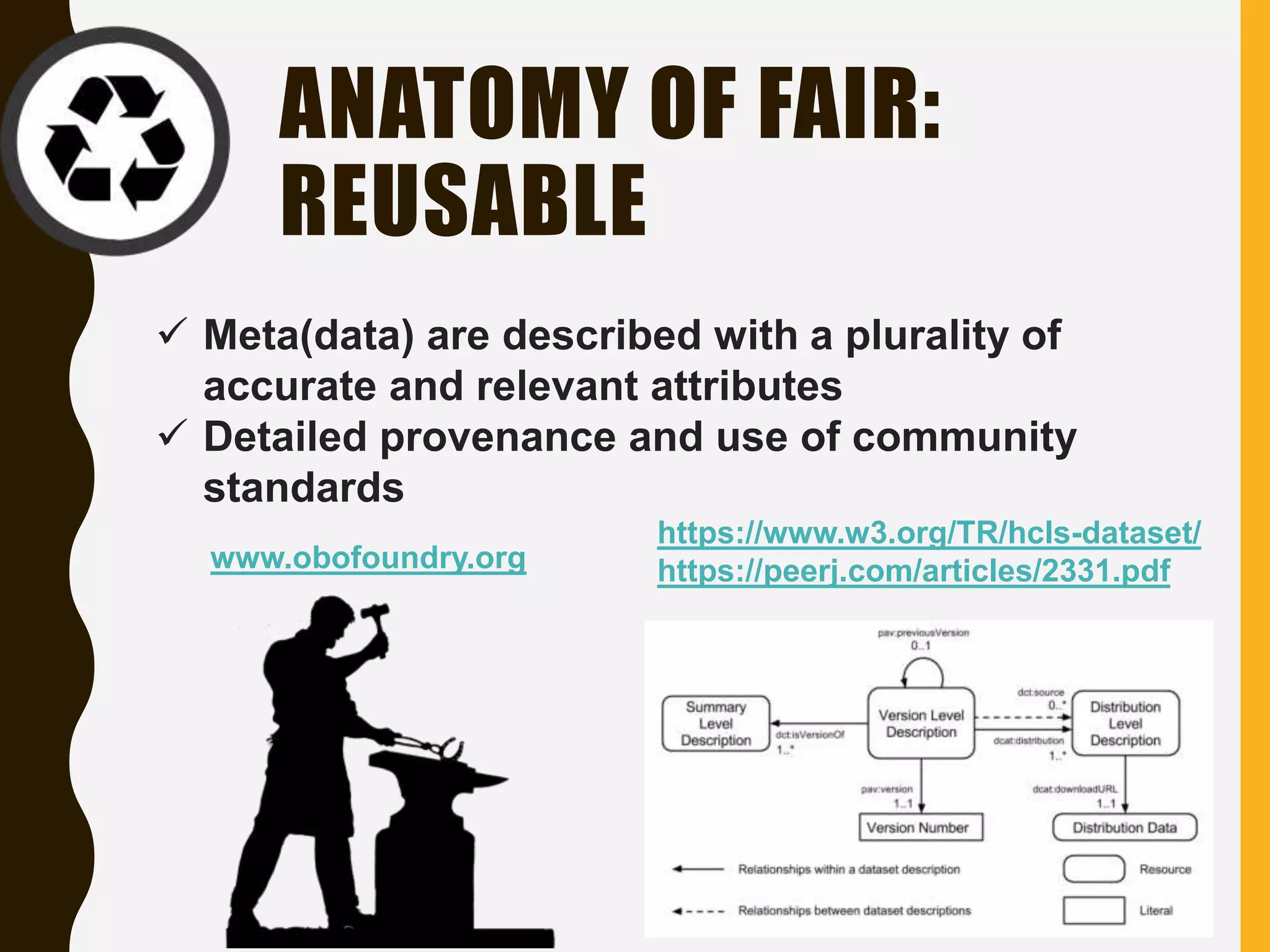
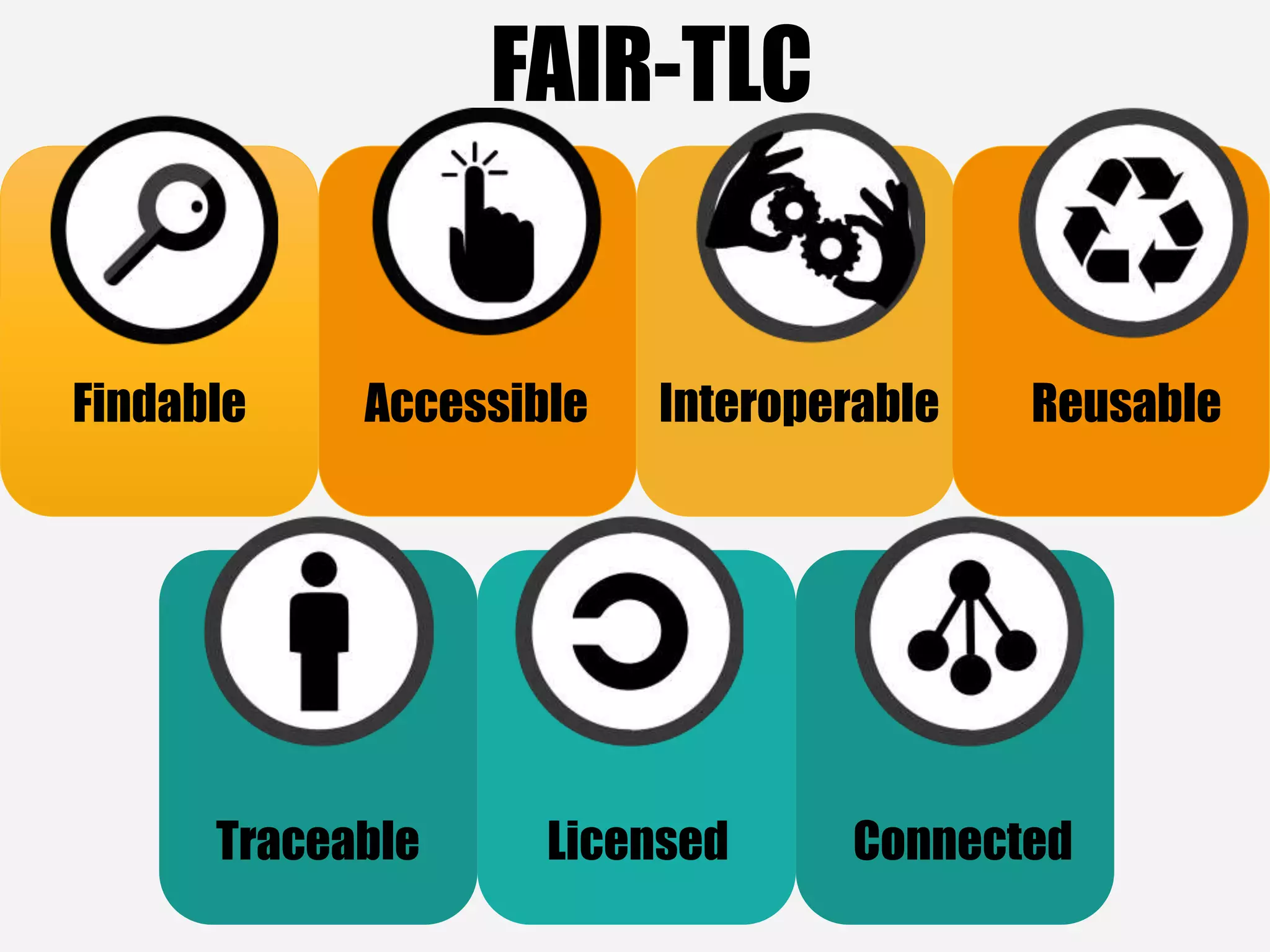
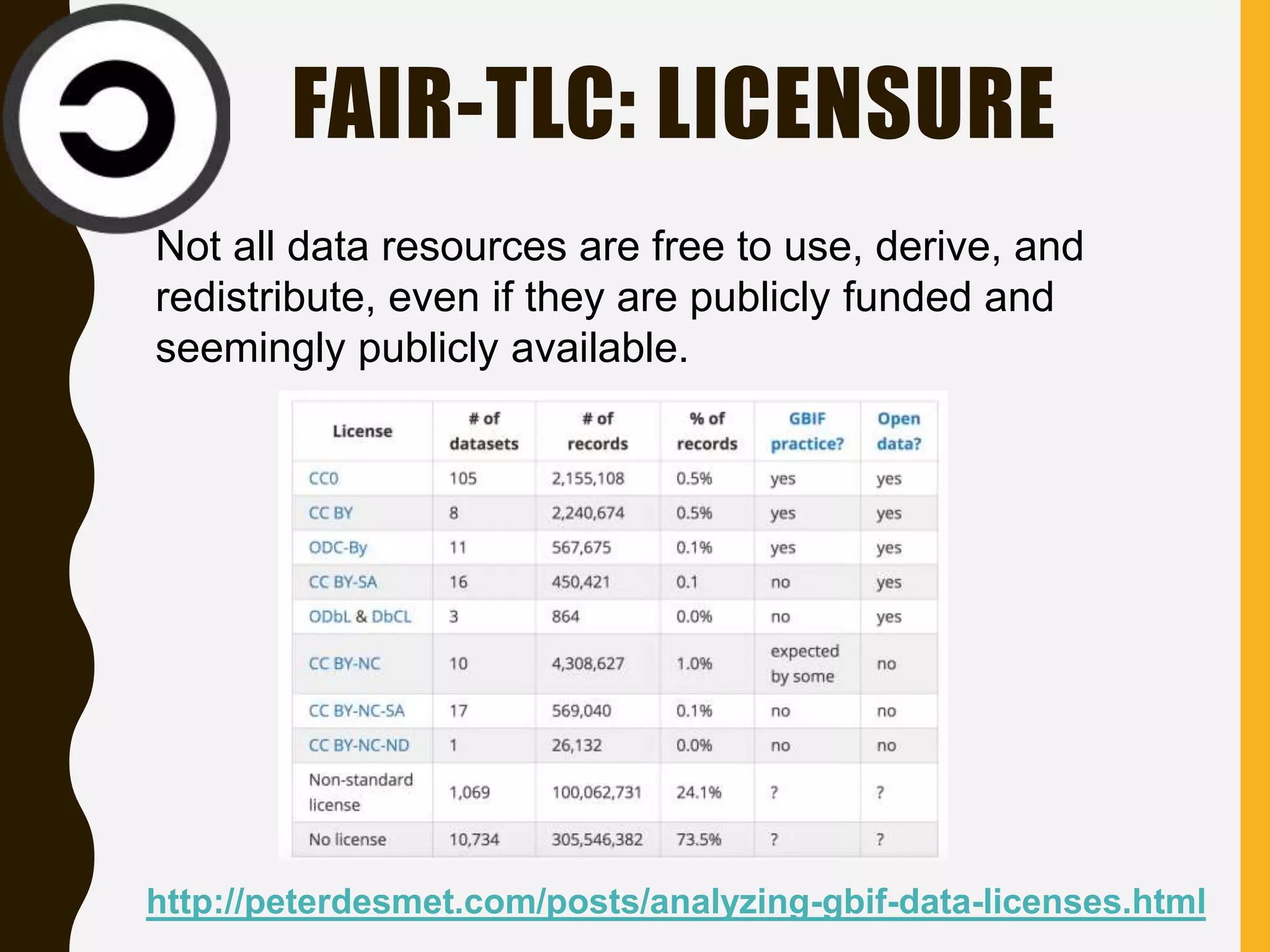
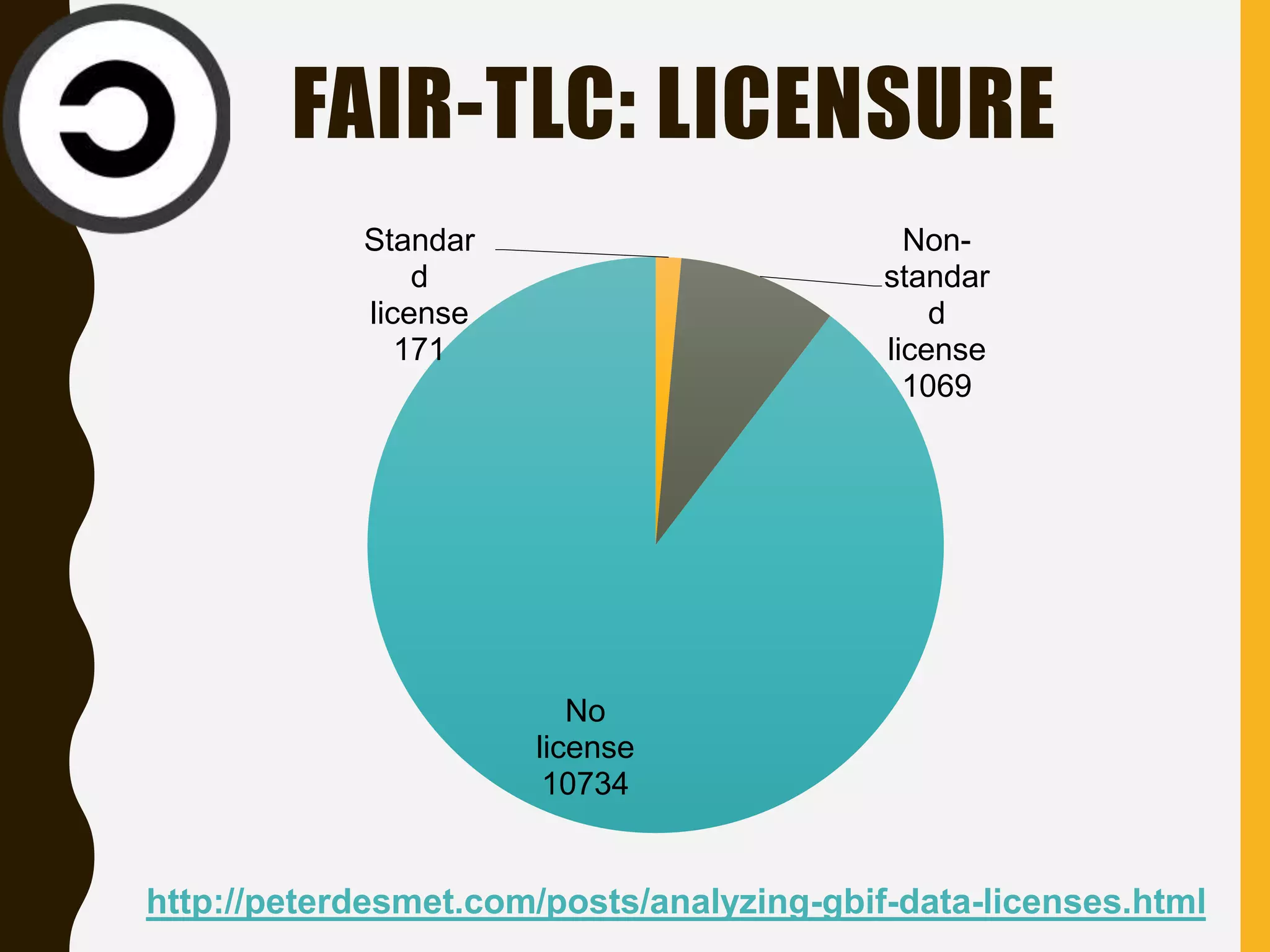
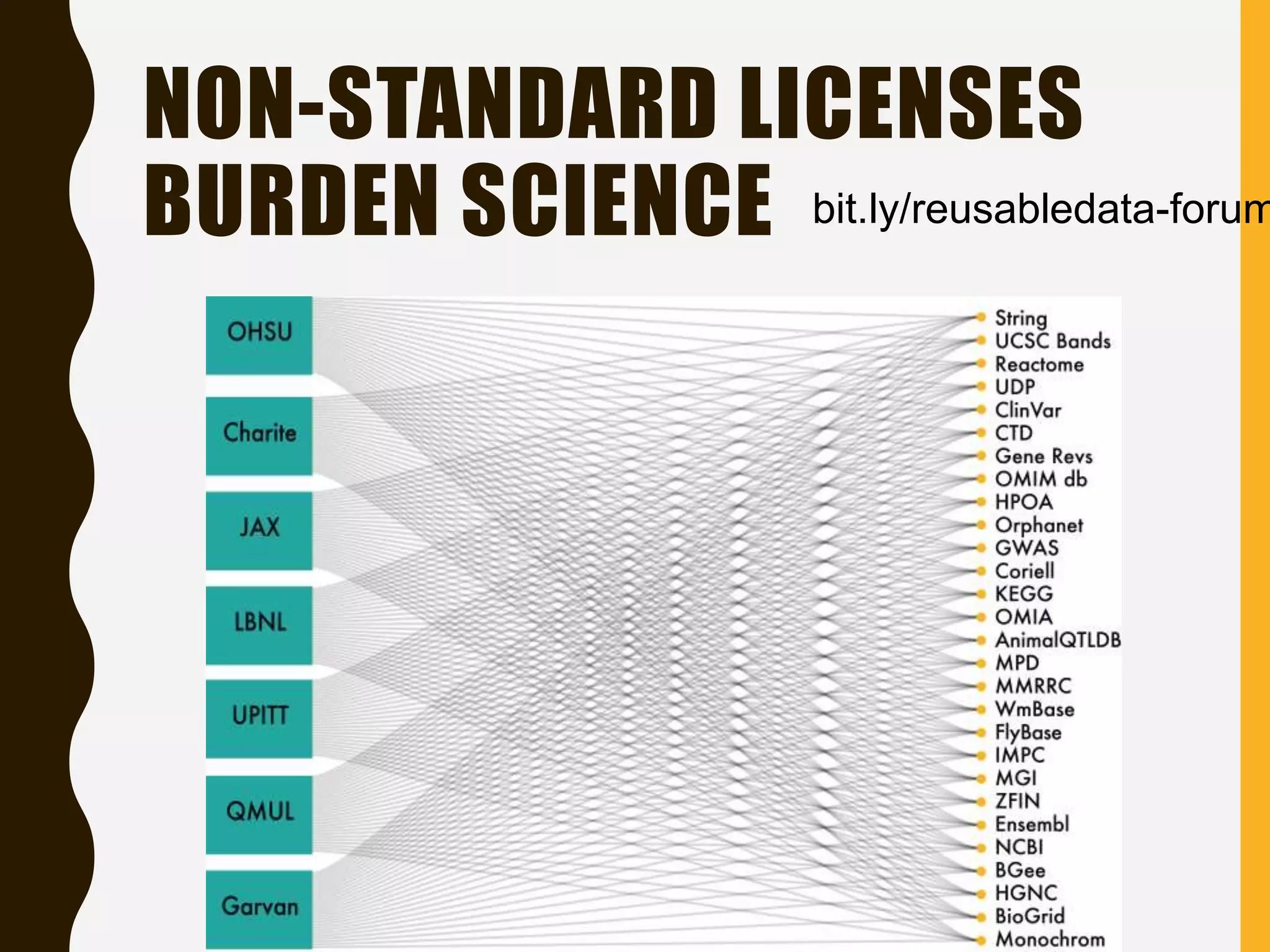
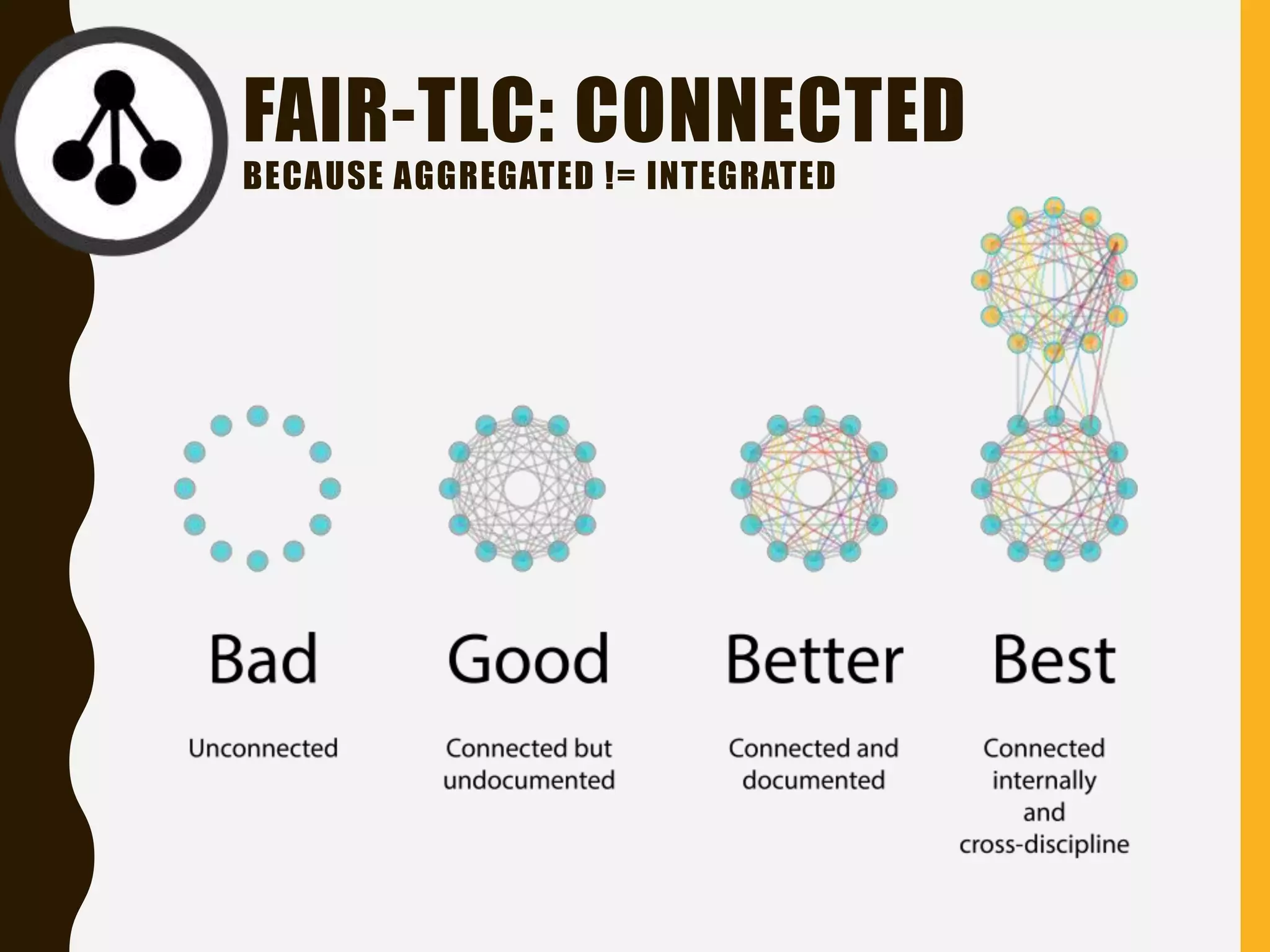
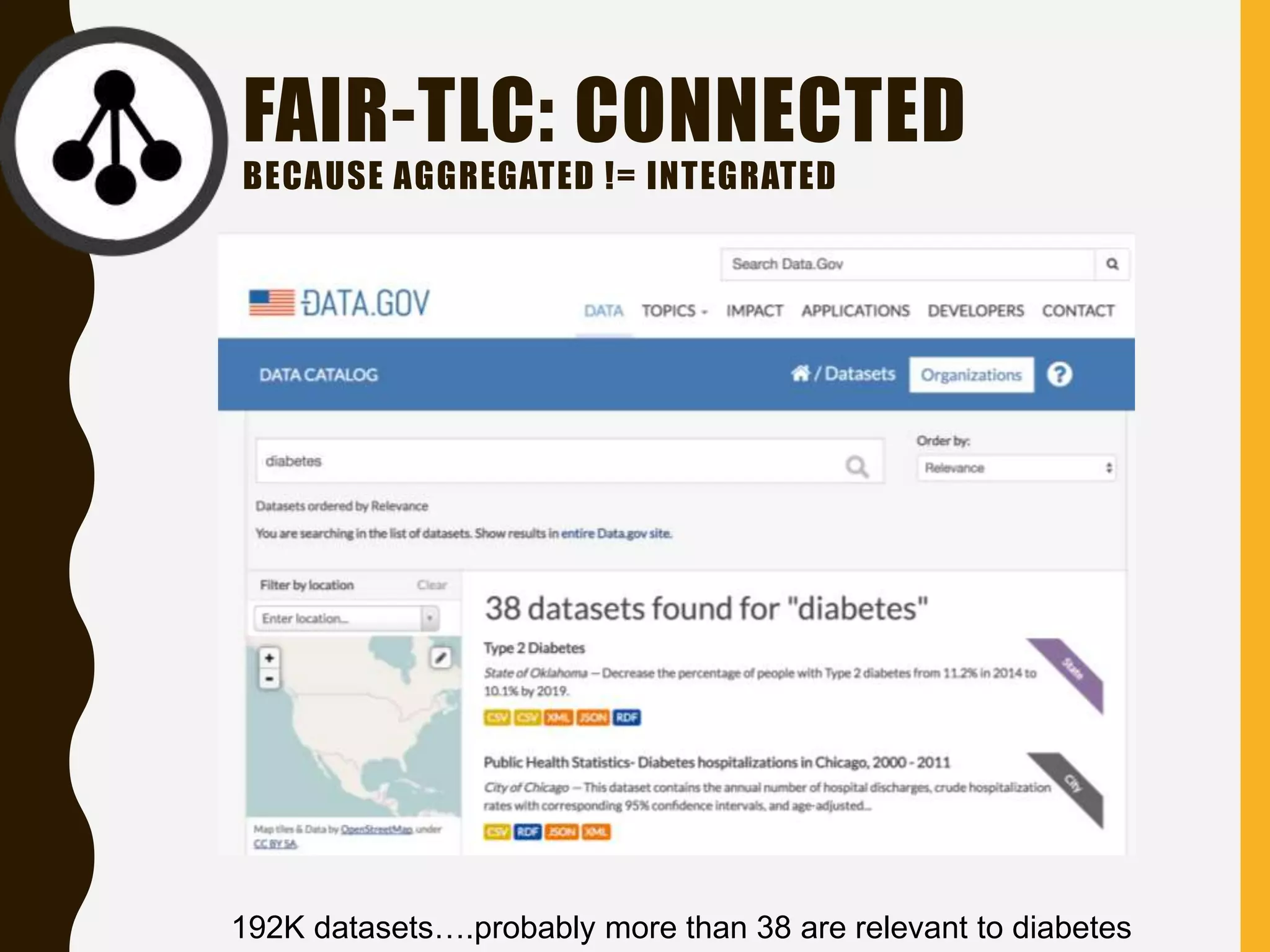
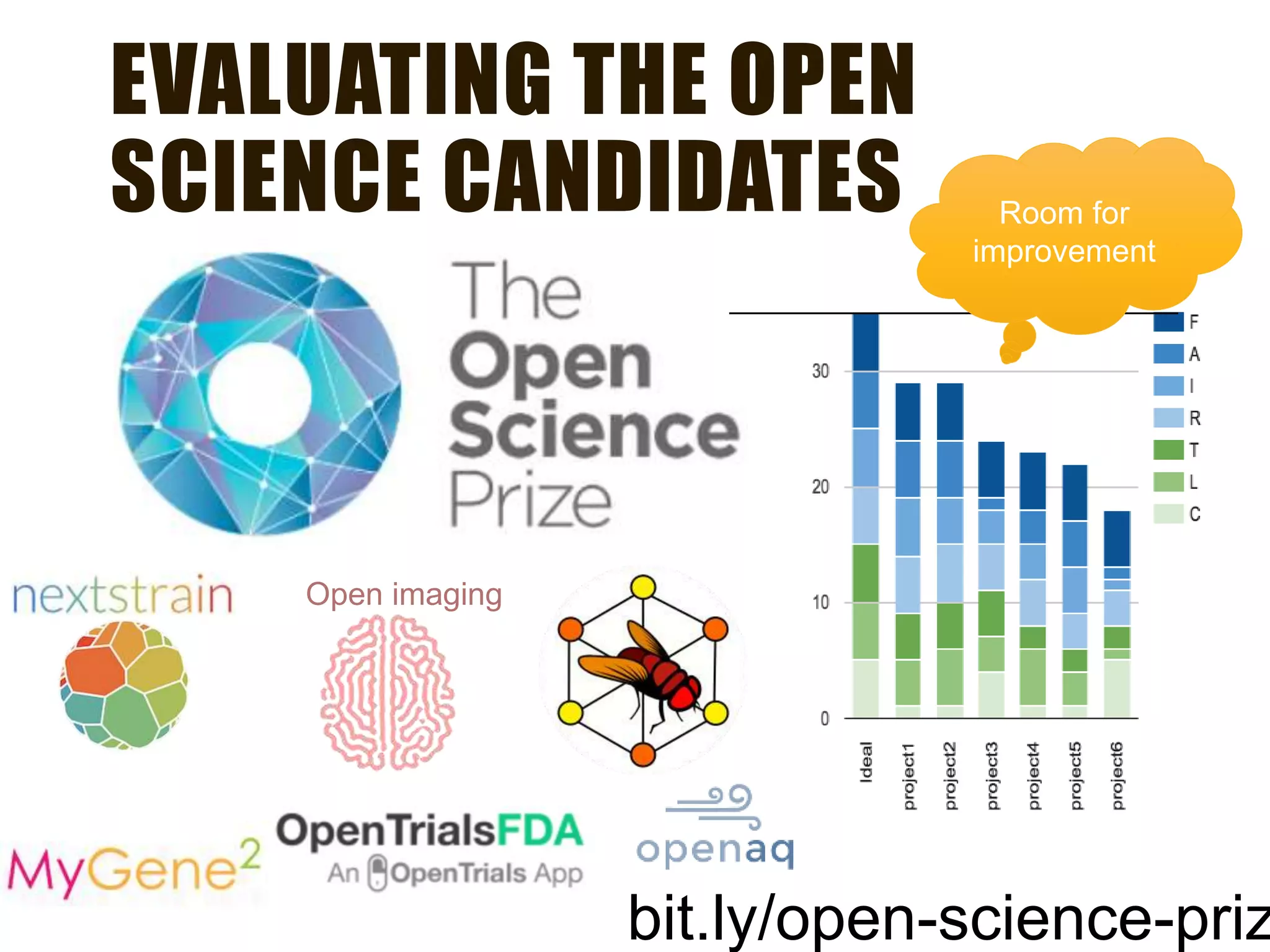
The document evaluates the openness of public knowledge bases, particularly in the context of the Nucleic Acids Research database collection, emphasizing the need for adherence to FAIR principles: Findable, Accessible, Interoperable, and Reusable. It discusses challenges in achieving true openness, such as varying data licenses and the importance of traceability and connectivity in data resources. The document proposes improvements for open science practices and highlights the role of evaluation rubrics and community standards in fostering better data sharing and usage.