The document presents a framework for evaluating and utilizing medical terminology mappings, highlighting the complexities and challenges faced in mapping terminologies across different use cases. It aims to enable collaborative semantics among providers and consumers of terminology mappings by inferring new mappings, presenting provenance, and performing validation. The framework incorporates various mapping strategies and emphasizes the importance of using RDF standards for representing mappings and their justifications.




























![Acknowledgments
• Session chair
• MIE2014 organizers
• SALUS team: Hong Sun, Ali Anil Sinaci, Gokce Banu Laleci Erturkmen
– Support from the European Community’s Seventh Framework Programme
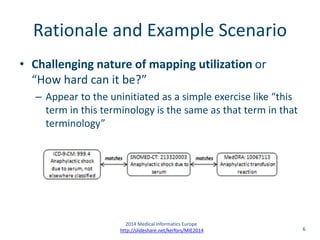
(FP7/2007–2013) under Grant Agreement No. ICT-287800, SALUS Project
(Scalable, Standard based Interoperability Framework for Sustainable
Proactive Post Market Safety Studies).
• EHR4CR team: WP4, WPG2, WP7 members
– Support from the Innovative Medicines Initiative Joint Undertaking under
grant agreement n° [No 115189]. European Union's Seventh Framework
Programme (FP7/2007-2013) and EFPIA companies
• Open PHACTS team: Alasdair Gray
• W3C HCLS team: Eric Prud’Hommeaux, Charlie Mead
31
2014 Medical Informatics Europe
http://slideshare.net/kerfors/MIE2014](https://image.slidesharecdn.com/mie2014-140825074303-phpapp02/85/MIE2014-A-Framework-for-Evaluating-and-Utilizing-Medical-Terminology-Mappings-31-320.jpg)






![Mapping Provenance Representation
38
:NanoPub_1_Supporting_2 = {
[ a r:Proof, r:Conjunction;
r:component <#lemma1>;
r:component <#lemma2>;
r:component <#lemma3>;
r:component <#lemma4>;
r:component <#lemma5>;
r:component <#lemma6>;
r:gives {
<http://purl.bioontology.org/ontology/ICD9CM/999.4> skos:prefLabel "Anaphylactic shock due to serum, not elsewhere classified".
<http://purl.bioontology.org/ontology/SNOMEDCT/213320003> skos:prefLabel "Anaphylactic shock due to serum".
<http://purl.bioontology.org/ontology/MDR/10067113> skos:prefLabel "Anaphylactic transfusion reaction".
<http://purl.bioontology.org/ontology/ICD9CM/999.4> skos:exactMatch <http://purl.bioontology.org/ontology/SNOMEDCT/213320003>.
<http://purl.bioontology.org/ontology/SNOMEDCT/213320003> skos:exactMatch <http://purl.bioontology.org/ontology/MDR/10067113>.
<http://purl.bioontology.org/ontology/ICD9CM/999.4> skos:exactMatch <http://purl.bioontology.org/ontology/MDR/10067113>.
}].
…….
…….
…….
<#lemma13> a r:Inference; r:gives {<http://purl.bioontology.org/ontology/ICD9CM/999.4> skos:exactMatch
<http://purl.bioontology.org/ontology/MDR/10067113>}; r:evidence (
<#lemma11>
<#lemma12>);
r:binding [ r:variable [ n3:uri "http://localhost/var#x0"]; r:boundTo [ n3:uri "http://purl.bioontology.org/ontology/ICD9CM/999.4"]];
r:binding [ r:variable [ n3:uri "http://localhost/var#x1"]; r:boundTo [ n3:uri "http://purl.bioontology.org/ontology/SNOMEDCT/213320003"]];
r:binding [ r:variable [ n3:uri "http://localhost/var#x2"]; r:boundTo [ n3:uri "http://purl.bioontology.org/ontology/MDR/10067113"]];
r:rule <#lemma14>.
<#lemma14> a r:Extraction; r:gives {@forAll var:x0, var:x1, var:x2. {var:x0 skos:exactMatch var:x1.
var:x1 skos:exactMatch var:x2} => {var:x0 skos:exactMatch var:x2}};
r:because [ a r:Parsing; r:source <file:///Users/sajjad/workspace/terminology-reasoning-test-case/example-term-map.n3>].
}.](https://image.slidesharecdn.com/mie2014-140825074303-phpapp02/85/MIE2014-A-Framework-for-Evaluating-and-Utilizing-Medical-Terminology-Mappings-38-320.jpg)