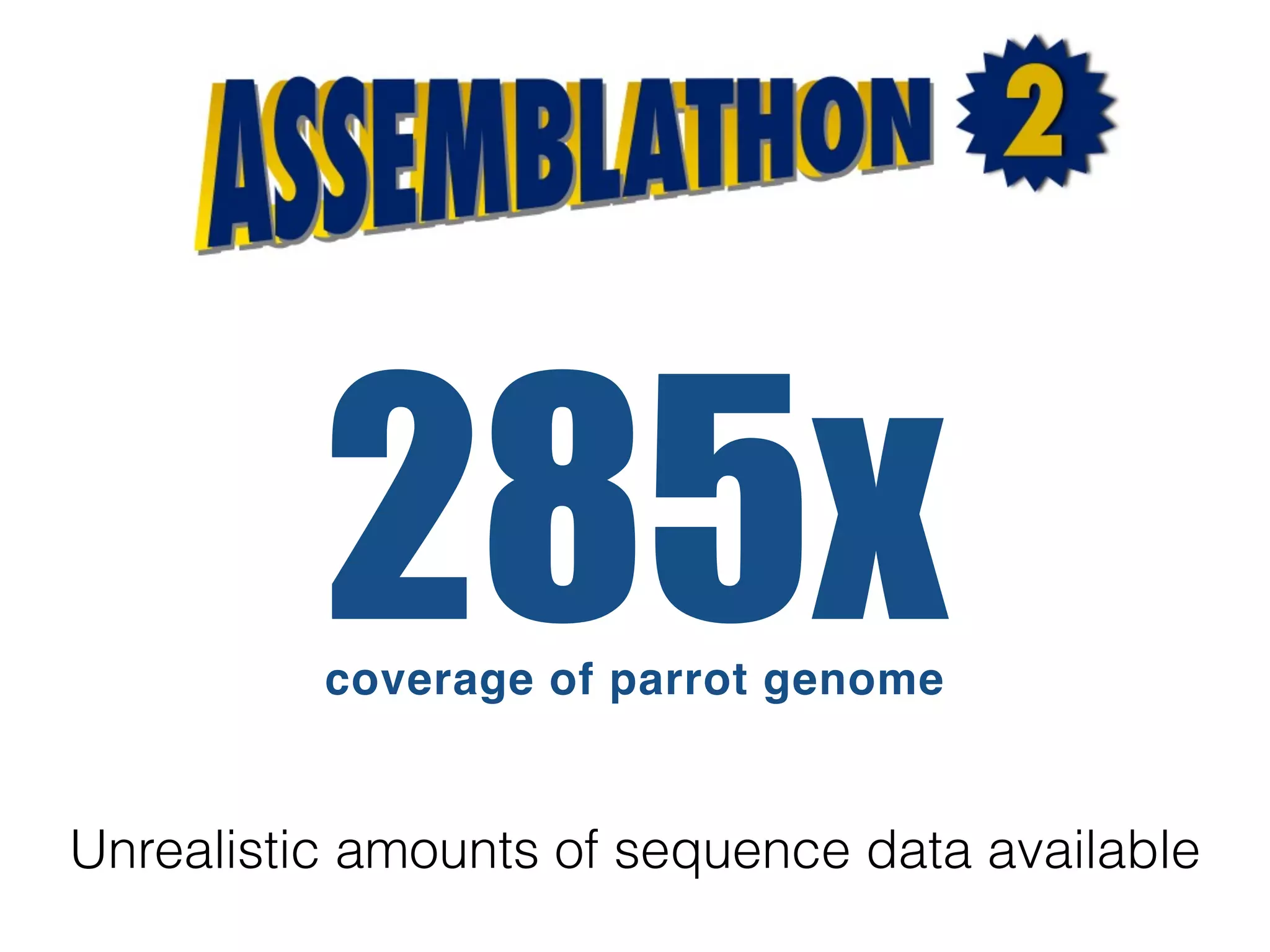
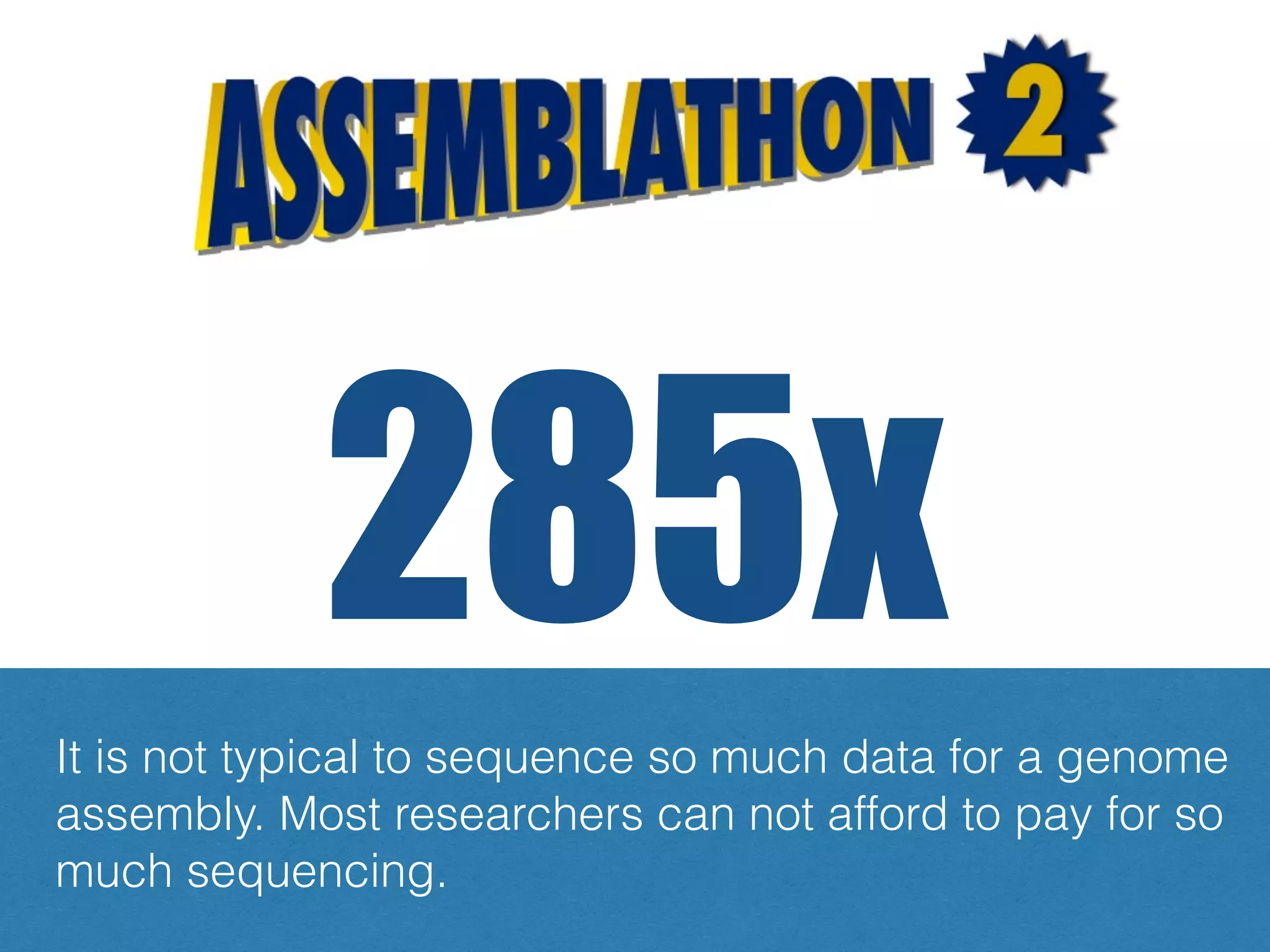
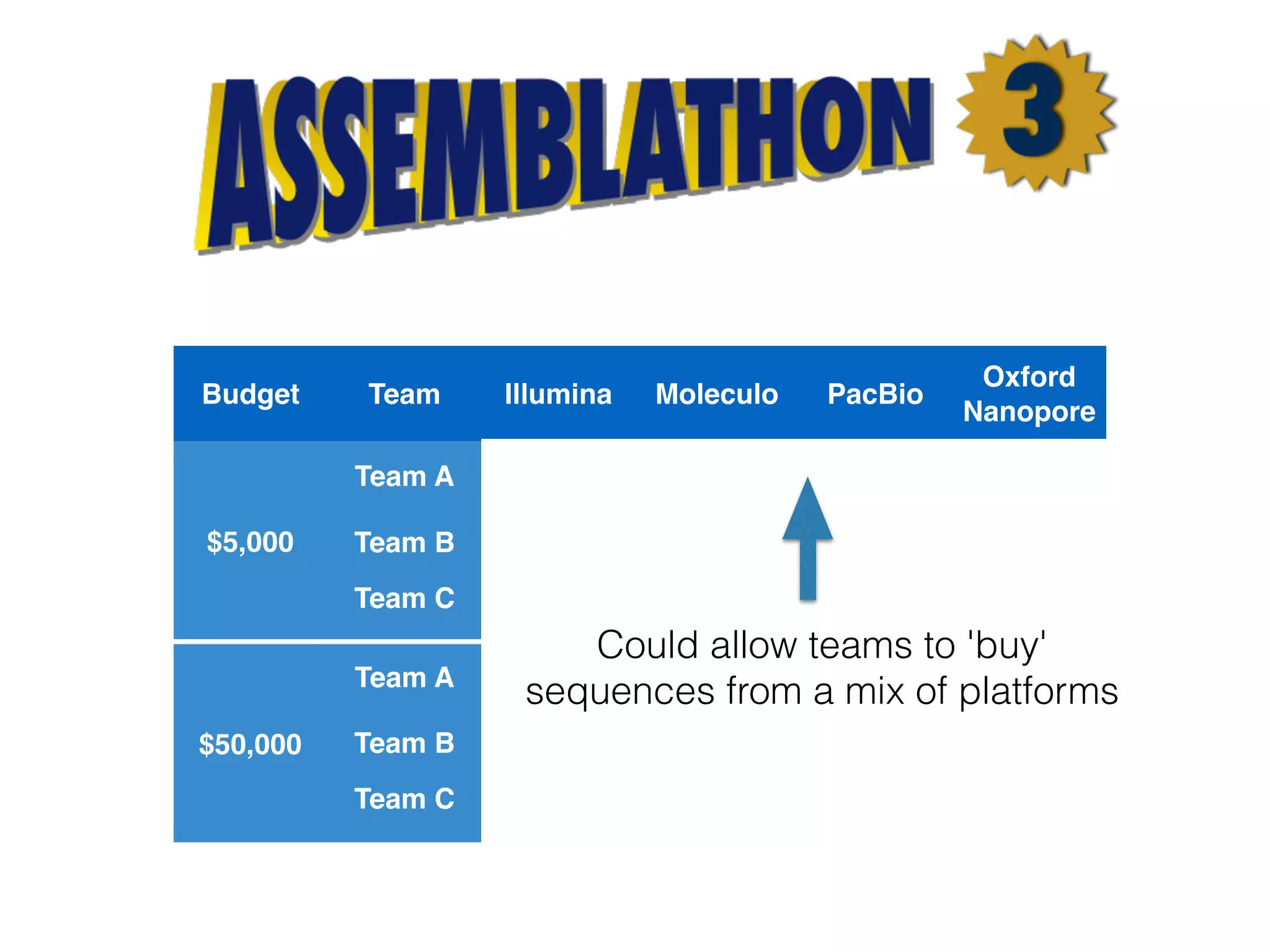
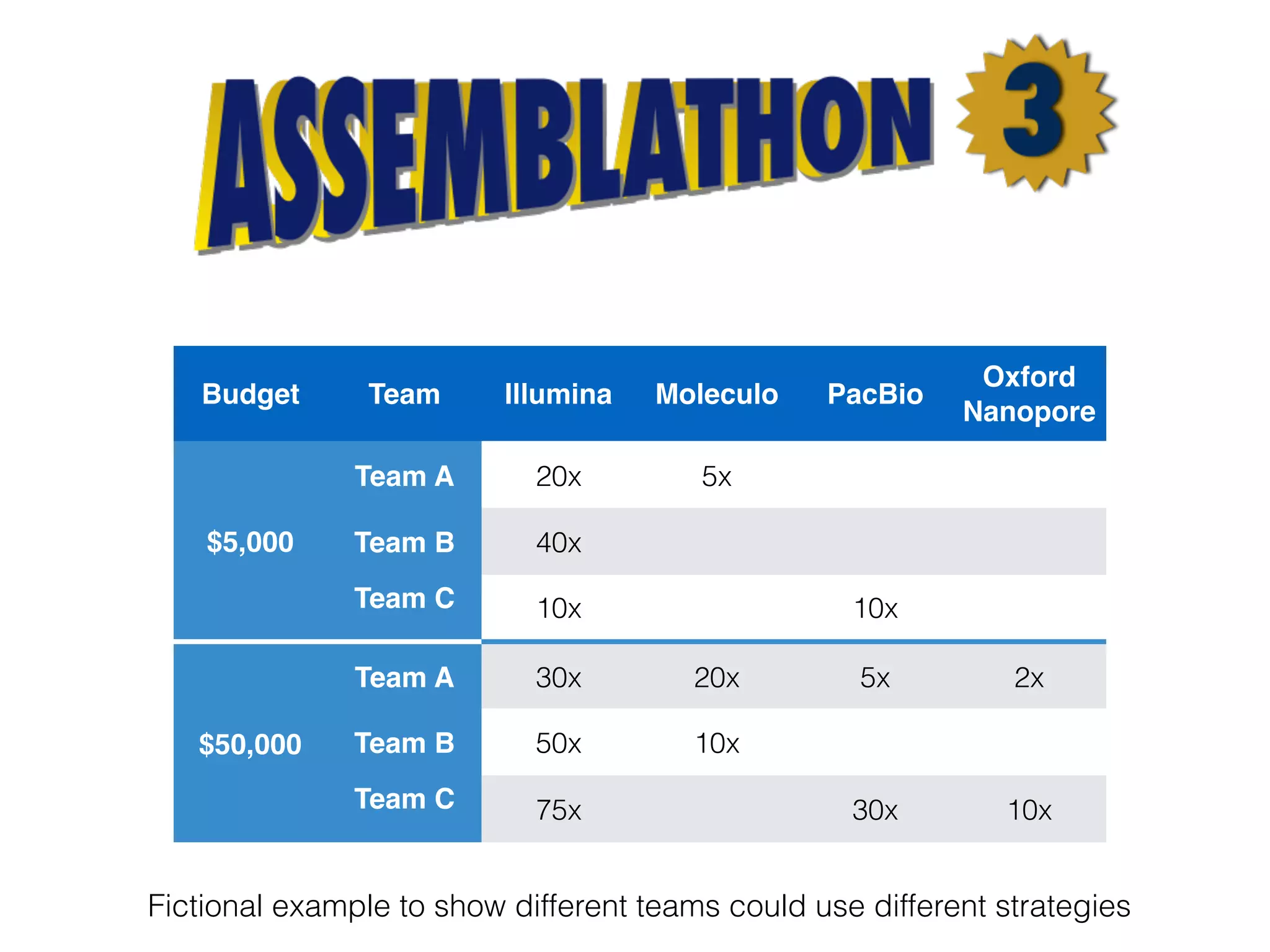
The document discusses ideas and challenges for a potential Assemblathon 3 event, highlighting the need for more practical genome assembly scenarios and community input for species selection. It critiques past Assemblathon experiences regarding data management, assembly validation, and technical specifications, emphasizing the importance of reproducible methodologies. Suggestions include focusing on endangered species or creating budget constraints to encourage realistic assembly strategies among teams.