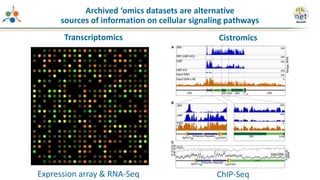
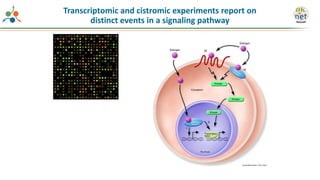
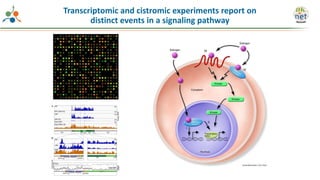
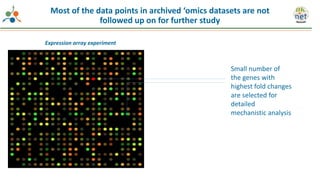
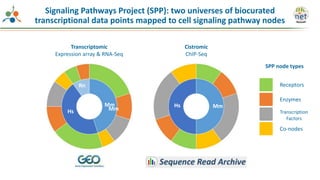
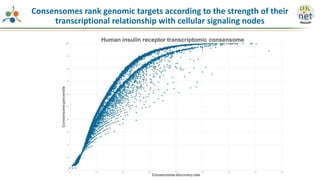
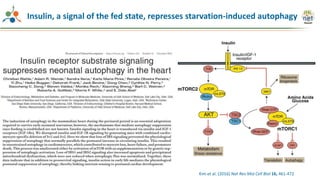
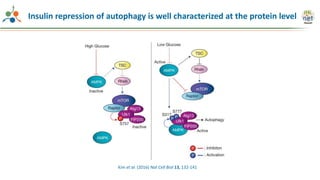
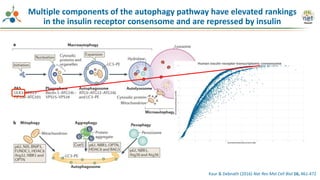
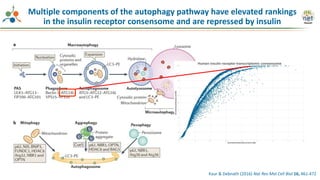
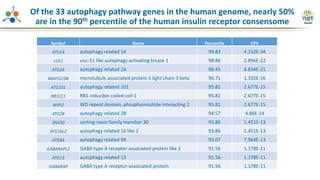
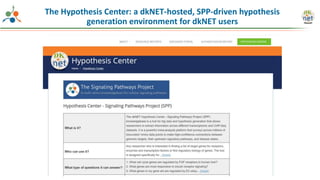
The Signaling Pathways Project (SPP) is an integrated knowledgebase focused on mammalian cellular signaling pathways, utilizing archived 'omics datasets to enhance accessibility and facilitate research. The project offers a user interface for mining data related to gene regulation, hypothesizing interactions between various signaling pathways and conditions, such as insulin secretion and Crohn's disease. Collaborative efforts aim to connect literature and 'omics data to drive discovery and understanding in cellular signaling.