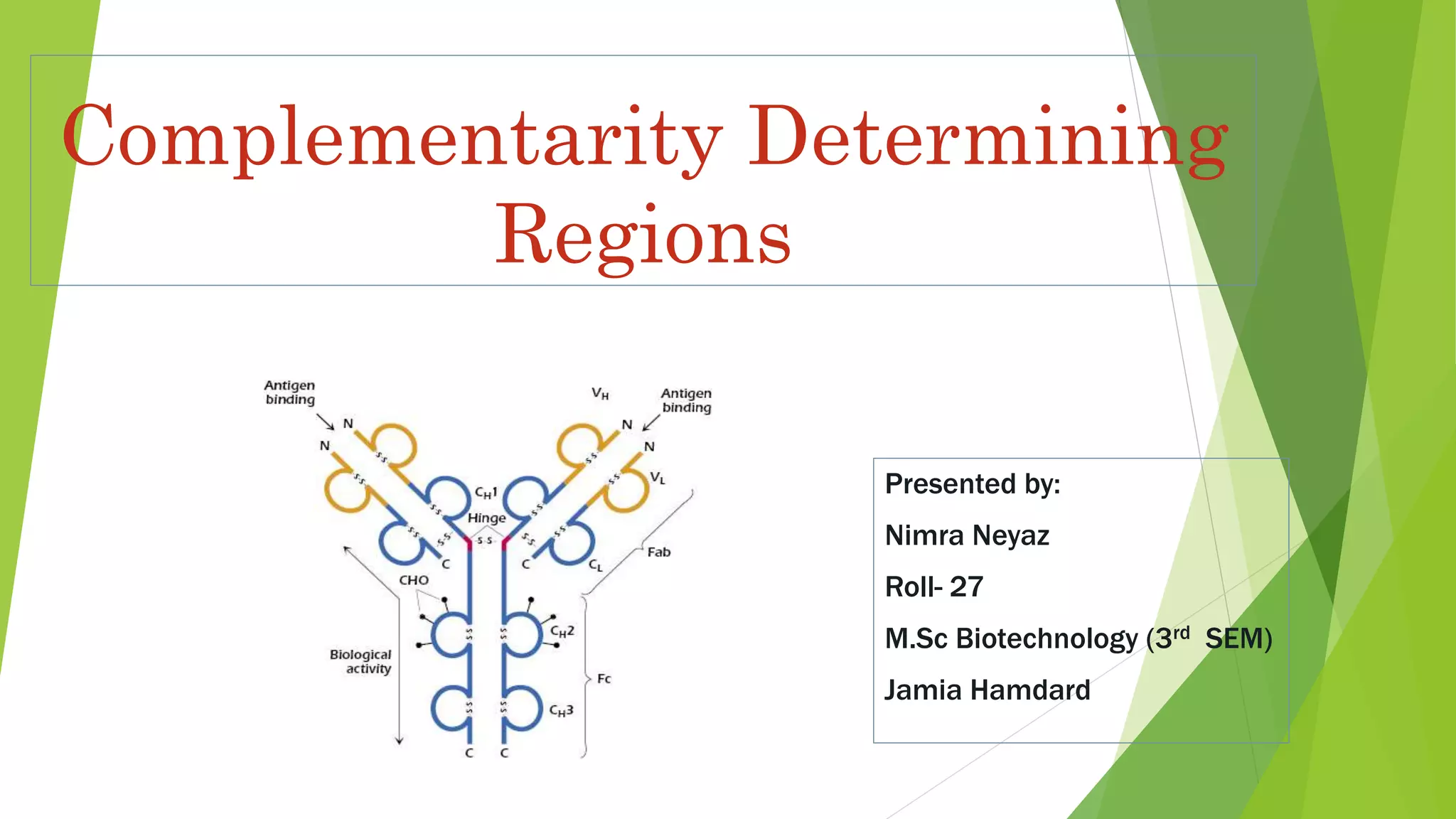
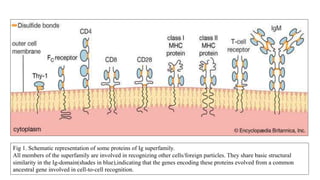
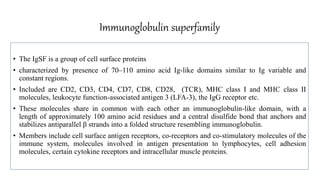
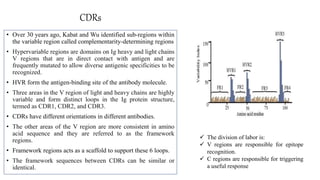
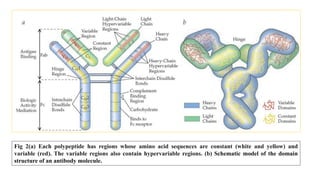
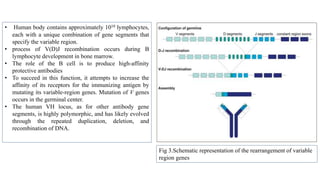
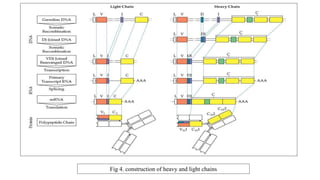
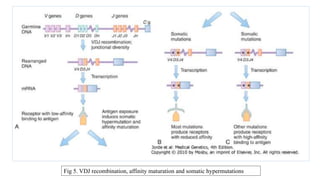
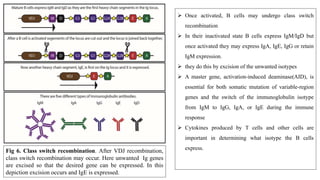
The document discusses the immunoglobulin superfamily (igsf) of proteins, which play crucial roles in immune system functions, particularly in cell recognition and antibody diversity. It details the structure of antibodies, emphasizing the significance of complementarity-determining regions (CDRs) in antigen binding, and the processes of V(D)J recombination, somatic hypermutation, and affinity maturation that enhance antibody specificity. Additionally, it describes how B cells undergo class switch recombination to express different immunoglobulin isotypes during an immune response.