Microbial eukaryotes in the Movile Cave chemosynthetic ecosystem
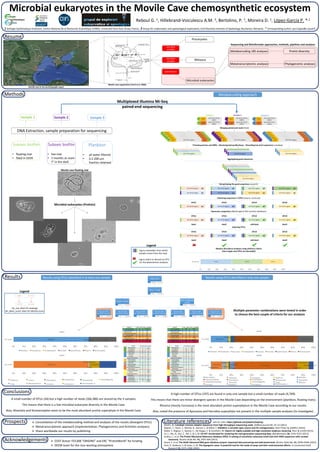
- 1. Methods Results Resume Microbial eukaryotes in the Movile Cave chemosynthetic ecosystem 1 Ecologie Systématique Evolution, Centre National de la Recherche Scientifique (CNRS), Université Paris-Sud, Orsay, France ; 2 Group for underwater and speleological exploration, Emil Racovita Institute of Speleology, Bucharest, Romania ; * Corresponding author: puri.lopez@u-psud.fr Prospects Literature references John St. John, https://github.com/jstjohn/SeqPrep Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ Prepr. 4, e2409v1 (2016). Mahé, F., Rognes, T., Quince, C., De Vargas, C. & Dunthorn, M. Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ 3, e1420 (2015). Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). Guillou, L. et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597-604 (2013). Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). Forti, P., Galdenzi, S. & Sarbu, S. M. The hypogenic caves: A powerful tool for the study of seeps and their environmental effects. in Continental Shelf Research 22, 2373–2386 (2002). Ø Consolidation of the metabarcoding method and analyses of the results (divergent OTUs) Ø Metatranscriptomic approach (Implementation, Phylogenomics and Activities analyses) Ø Share worldwide our results by publishing Reboul G. 1, Hillebrand-Voiculescu A.M. 2, Bertolino, P. 1, Moreira D. 1, López-García P. *,1 Metabarcoding approach Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID Forward primer Reverse primer 18 S V4-V5 regionsMID MID 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions Trimming primers and MIDs - Removing bad quality bases - Discarding too short sequences (cutadapt) Reverse primer part of 18 S V4-V5 regions MID part of 18 S V4-V5 regions MID Forward primer Reverse primer part of 18 S V4-V5 regions MID part of 18 S V4-V5 regions MID Forward primer Reverse primer part of 18 S V4-V5 regions MID part of 18 S V4-V5 regions MID Forward primer Merging paired-end reads (flash) 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regionsx1 x1 x2 x1 Dereplicating the good sequences (vsearch) 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions x1x1x1x1 Clustering sequences in OTUs (Swarm, cd-hit-est) 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions x2x3x3x1 OTU1 OTU2 OTU3 OTU4 Taxonomic assignation (BlastN against PR2 and Silva databases) Aggregating good sequences 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions x2x3x3x1 OTU1 OTU2 OTU3 OTU4 taxa1 taxa2 unknown taxa3 Selecting OTUs 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions 18 S V4-V5 regions x2x3x3x1 OTU1 OTU2 OTU3 OTU4 taxa1 taxa2 unknown taxa3 OTU2 OTU3 OTU4Abundance 0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% Relative abundance analyses using arbitrary criteria (here single read OTUs are discarded) Sample 1 Sample 2 Sample 3 Alveolata Opisthokonta Stramenopiles 0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% tot_reads rank1 Alveolata Amoebozoa Archaeplastida Excavata Opisthokonta Rhizaria Stramenopiles Ciliophora Choanoflagellida Fungi Ochrophyta Stramenopiles_X 0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% tot_reads rank2 Ciliophora Conosa Chlorophyta Streptophyta Metamonada Choanoflagellida Fungi Mesomycetozoa Cercozoa Ochrophyta Stramenopiles_X Alveolata Archaeplastida Hacrobia Opisthokonta Rhizaria Stramenopiles 0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% tot_reads rank1 Alveolata Amoebozoa Apusozoa Archaeplastida Excavata Hacrobia Opisthokonta Rhizaria Stramenopiles Apicomplexa Ciliophora Dinophyta Perkinsea Chlorophyta Fungi Cercozoa Radiolaria Ochrophyta Stramenopiles_X 0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% tot_reads rank2 Apicomplexa Ciliophora Dinophyta Perkinsea Breviatea Apusomonadidae Chlorophyta Streptophyta Metamonada Haptophyta Katablepharidophyta Telonemia Choanoflagellida Fungi Metazoa Cercozoa Radiolaria Ochrophyta Stramenopiles_X CDhit OTUs 8,488 142,371 OTUs > 1 read 301 134,184 multiple samples 37 113,883 bh_cov > 70 bh_ident_score>80 34 106,386 bh_cov > 80 bh_ident_score>80 33 105,852 bh_cov > 90 bh_ident_score>80 30 88,418 OTUs in only one sample 264 20,301 bh_cov > 80 bh_ident_score>80 238 18,885 bh_cov > 80 bh_ident_score>90 215 17,849 bh_cov > 90 bh_ident_score>80 207 4,793 bh_cov > 90 bh_ident_score>90 197 4,769 Acknowledgements Ø COST Action TD1308 “ORIGINS” and ERC “ProtistWorld” for funding Ø DEEM team for the nice working atmosphere Results using OTUs identified in at least one sample Results using OTUs identified in only one sample 8,488 142,371 nb OTUs nb reads Legend bh_cov: blast hit coverage bh_ident_score: blast hit identity score Multiplexed Illumina Mi-Seq paired-end sequencing Conclusions A small number of OTUs (34) but a high number of reads (106,386) are shared by the 3 samples. This means that there is a low microbial eukaryote diversity in the Movile Cave. Also, Alveolata and Stramenopiles seem to be the most abundant protist superphyla in the Movile Cave. Movile cave organization (Forti et al. 2002) Movile cave in the world (google maps) Microbial eukaryotes Metazoa Procaryotes HAS BEEN STUDIED HAS BEEN STUDIED OUR PROJECT Protist diversityMetabarcoding 18S analyses Phylogenomic analysesMetatranscriptomic analyses Sequencing and Bioinformatic approaches, methods, pipelines and analyses A high number of OTUs (197) are found in only one sample but a small number of reads (4,769). This means that there are minor divergent species in the Movile Cave depending on the environment (plankton, floating mats). Rhizaria (mostly Cercozoa) is the most abundant protist superphylum in the Movile Cave according to our results. Also, noted the presence of Apusozoa and Hacrobia superphyla not present in the multiple sample analyses (to investigate). Multiple parameter combinations were tested in order to choose the best couple of criteria for our analyses Sample 1 Sample 2 Sample 3 Suboxic biofilm • floating mat • fixed in EtOH Suboxic biofilm • live mat • 2 months at room T° in the dark Plankton • all water filtered • 0.2-200 µm fraction retained DNA Extraction, sample preparation for sequencing Legend : tag to remember from which sample comes from the read : tag to select or discard an OTU for the downstream analyses Movile cave floating mat Microbial eukaryotes (Protists)