MIT Amgen Scholar Presentation
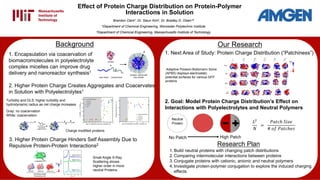
- 1. Effect of Protein Charge Distribution on Protein-Polymer Interactions in Solution Brandon Clark†, Dr. Sieun Kim‡, Dr. Bradley D. Olsen*‡ †Department of Chemical Engineering, Worcester Polytechnic Institute ‡Department of Chemical Engineering, Massachusetts Institute of Technology Background Our Research Research Plan 1. Build neutral proteins with changing patch distributions 2. Comparing intermolecular interactions between proteins 3. Conjugate proteins with cationic, anionic and neutral polymers 4. Investigate protein-polymer conjugation to explore the induced charging effects 1. Next Area of Study: Protein Charge Distribution (“Patchiness”) 3. Higher Protein Charge Hinders Self Assembly Due to Repulsive Protein-Protein Interactions2 Small Angle X-Ray Scattering shows higher order in more neutral Proteins 2. Goal: Model Protein Charge Distribution’s Effect on Interactions with Polyelectrolytes and Neutral Polymers 𝐿2 𝑁 = 𝑃𝑎𝑡𝑐ℎ 𝑆𝑖𝑧𝑒 # 𝑜𝑓 𝑃𝑎𝑡𝑐ℎ𝑒𝑠 Neutral Protein No Patch High Patch Adaptive Poisson-Boltzmann Solve (APBS) displays electrostatic potential surfaces for various GFP proteins 1. Encapsulation via coacervation of biomacromolecules in polyelectrolyte complex micelles can improve drug delivery and nanoreactor synthesis1 2. Higher Protein Charge Creates Aggregates and Coacervates in Solution with Polyelectrolytes1 Turbidity and DLS: higher turbidity and hydrodynamic radius as net charge increases Gray: no coacervation White: coacervation Charge modified proteins
- 2. 2. AIBN: radical source RAFT Polymerization Step 1: Step 2: 3. Chain Transfer Agent3 Maleimide-CTACTA Methods: Polymerization Polymerization for BioconjugationPolymerization for Separated Solutions M1 (Neutral) M2 (Cation) M3 (Anion) 1. Monomers Reactants Polyelectrolyte Formation Quaternized PDMAEMA: strong polycation PMAA: weak polyanionOriginal PDMAEMA: weak polycation Most likely too strong to compare with PMAA due to natural positive charge out of solution
- 3. Methods: Protein Synthesis Bacterial Transformation Lysed Cell Ncol Green Fluorescent Protein Hind3 Bioconjugation: Maleimide Thiol “Click” Reaction Bioconjugate Solutions 1. IPTG 2. Cell Lysis Ni-NTA column Ni-NTA 6xHis-tag On Proteins Protein Purification Bound Protein Eluting protein using Imidazol Filter Solutions of separated protein and polymer Advantages of this click reaction 1.Site specific 2.No catalyst 3.Water soluble 4.Quick reaction
- 4. Bioconjugation with PNIPAMPolymerization 3. GPC Shows Polymerization of Monomers for Separated Solutions Results 1. NMR for Chain Transfer Agents 1. SDS Gel and LC/MS confirms Protein Yield and Bioconjugation Maleimide-Functionalized PNIPAM MW: 48 kDa Bioconjugates Pure Proteins (-20) (0) J HP NP HPR 2. Proteins Retain Glow before and after Bioconjugation 300 400 500 600 700 800 0 20 40 60 80 100 523 485 Intensity m/z 507 [M+H] [M+Na] [M+K] 200 300 400 500 600 700 800 0 20 40 60 80 100 m/z [M+Na] 247 1-Propanol Maleimide Anhydride Mw = 224 Da Maleimide Functionalized Benzo-CTA Mw = 484 Da 2. LC-MS Mass Spectra for RAFT CTA Expected molar masses were seen using LC-MS. This indicates presence of desired compounds to use in RAFT reactions. LC/MS Molar Mass: 76kDa Protein MW: 28 kDa 0 5 10 15 20 25 30 -0.00004 -0.00002 0.00000 0.00002 0.00004 0.00006 0.00008 0.00010 0.00012 Retention time (min) 5 10 15 20 25 30 -0.00004 -0.00002 0.00000 0.00002 0.00004 Retention time (min) 0 5 10 15 20 25 30 -0.00002 0.00000 0.00002 0.00004 0.00006 0.00008 0.00010 0.00012 0.00014 Retention time (min) • PEG • MW: 33 kDa • PDI: 1.01 • PMAA • MW: 54 kDa? • PDI: 1.15 • PDMEAMA • MW: 26 kDa • PDI: 1.01 UV lamp at 365nm Since GFP typically glows when it is correctly folded, this indicates that both pure protein and bioconjugate samples contain correctly folded GFP. For bioconjugate solutions For separated solutions
- 5. Results, Conclusions, and Future Work AcknowledgmentsReferences This work was sponsored by the Amgen Foundation and facilitated by the Massachusetts Institute of Technology UROP Program. I would like to thank Amgen, MIT, Dr. Bradley Olsen, Dr. Sieun Kim, and the rest of the Olsen Lab for their support and mentorship. 1. Obermeyer, Allie C., et al. "Complex coacervation of supercharged proteins with polyelectrolytes." Soft matter 12.15 (2016): 3570-3581. 2. Lam, Christopher N., Helen Yao, and Bradley D. Olsen. "The Effect of Protein Electrostatic Interactions on Globular Protein–Polymer Block Copolymer Self-Assembly." Biomacromolecules 17.9 (2016): 2820-2829. 3. Chang, Dongsook, et al. "Effect of polymer chemistry on globular protein–polymer block copolymer self- assembly." Polymer Chemistry 5.17 (2014): 4884-4895. Turbidity Results Future Work 1. Successfully expressed GFP proteins with varied charge distributions 2. Various CTAs for RAFT polymerization were synthesized 3. Polymerized model polymers by RAFT polymerization with low polydispersity (PDI: ~1.1) 4. Bioconjugated mutated GFP proteins with PNIPAM, showing expected molecular weight and preserved protein folding 5. This study will show the effects of protein charge patch size on various intermolecular interactions DLS1 Different patch size of proteins may show various electrostatic interactions between molecules Turbidity can elucidate the extent that proteins form aggregates or coacervates with polymers and polyelectrolytes 100 80 60 40 20 0 0.0 0.2 0.4 0.6 0.8 1.0 Absat750nm Weight fraction, PEG C(-20) Janus Homogeneous 100 80 60 40 20 0 0.0 0.2 0.4 0.6 0.8 1.0 Absat750nm Weight fraction, qPDMAEMA C(-20) Janus Homogeneous 100 80 60 40 20 0 0.0 0.2 0.4 0.6 0.8 1.0 Absat750nm Weight fraction, PAA C(-20) Janus Homogeneous Preliminary data: GFP (-20) and Janus GFP show high turbidity in solution with the strong polycation Neutral: PEG Cationic: qPDMAEMA Anionic: PAA Protein Modeling Conclusions
