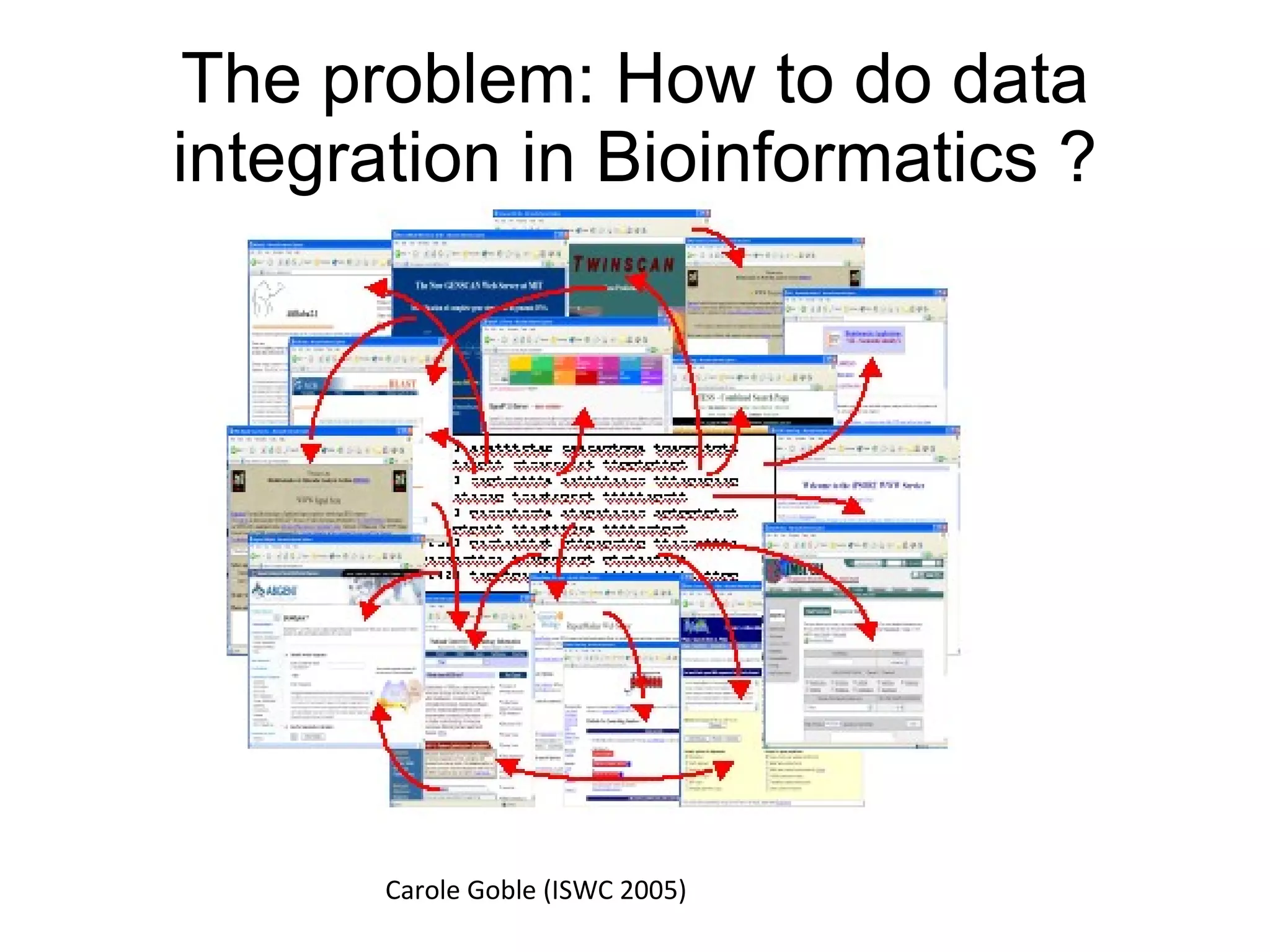
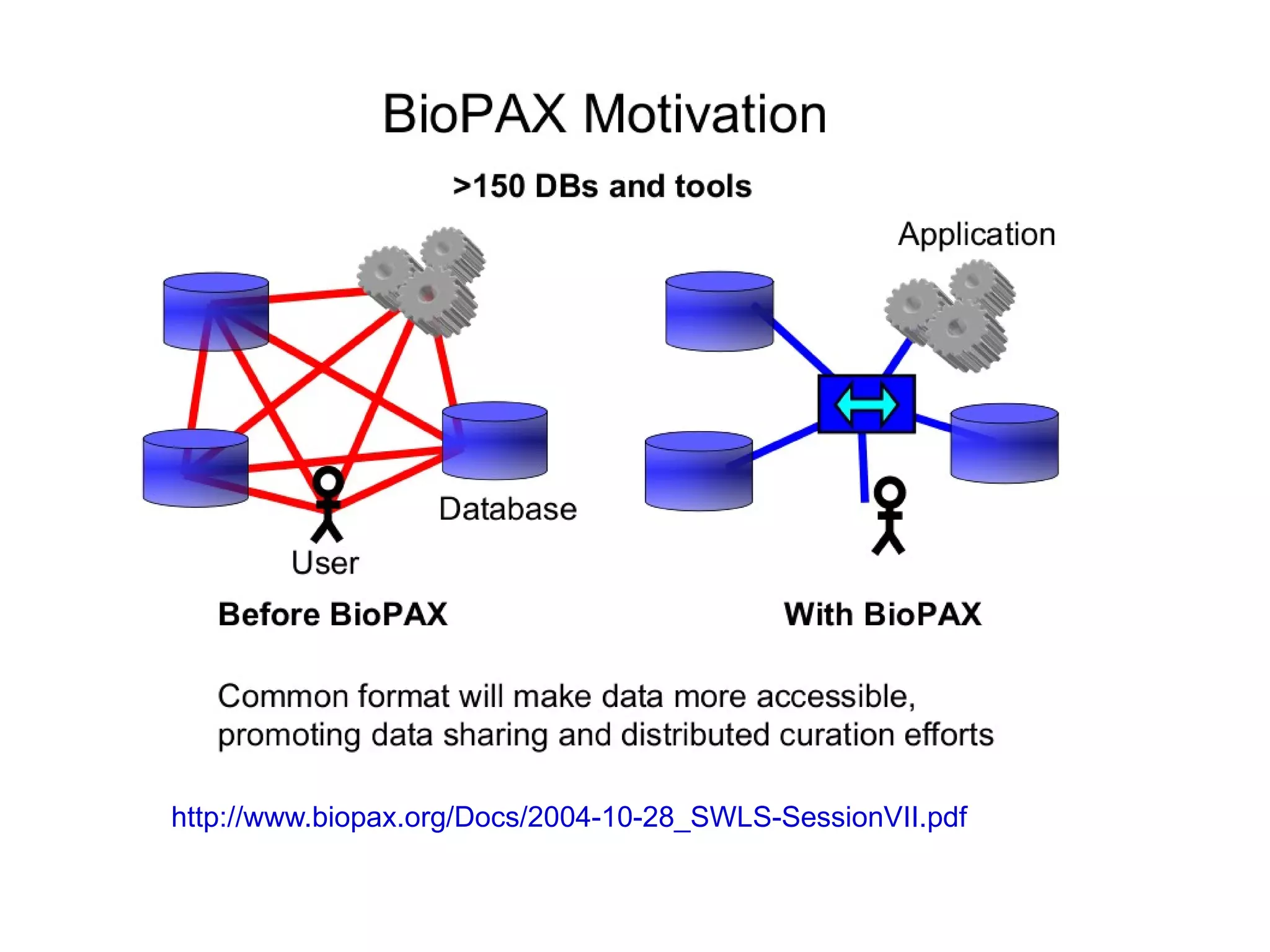
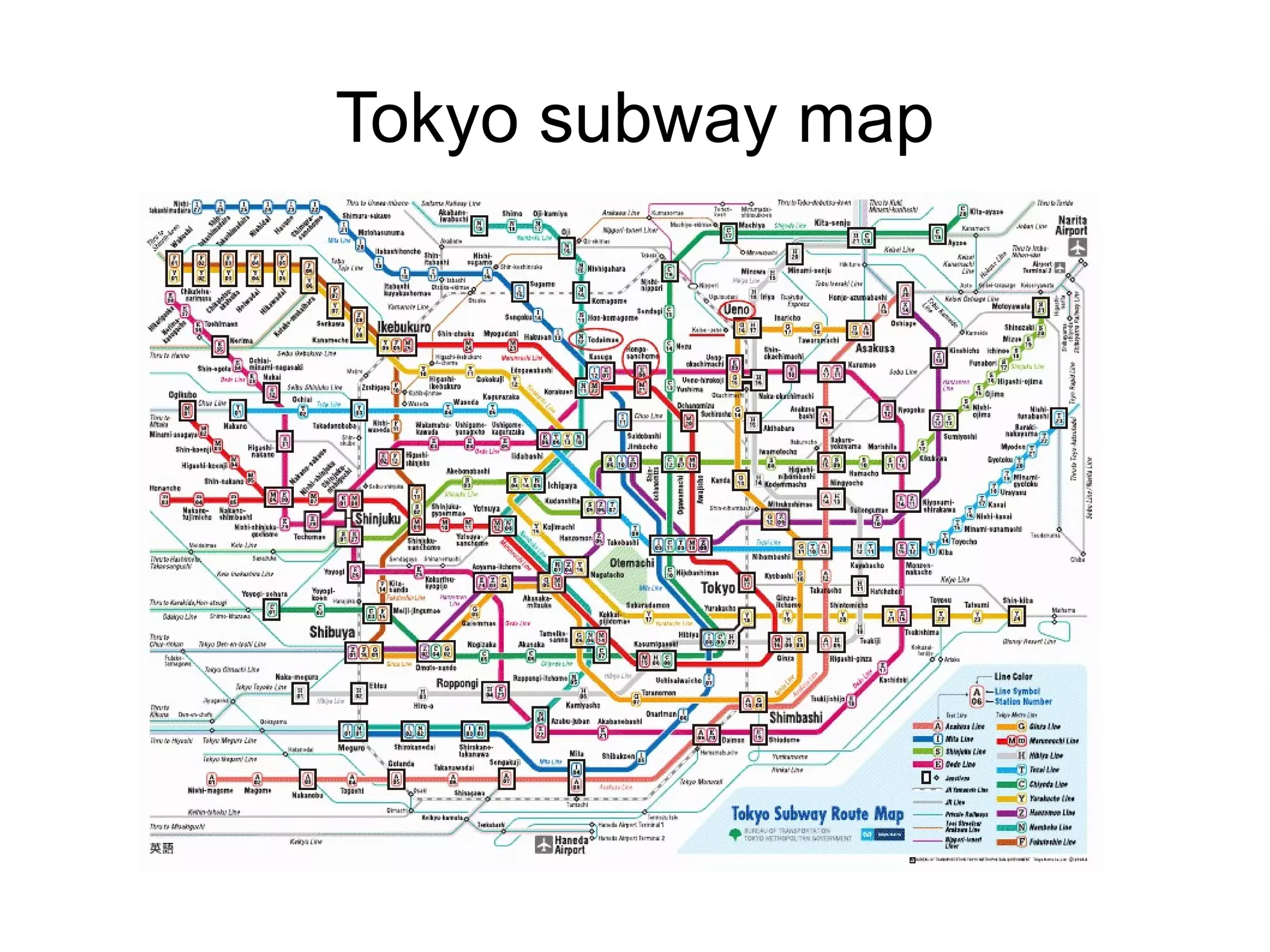
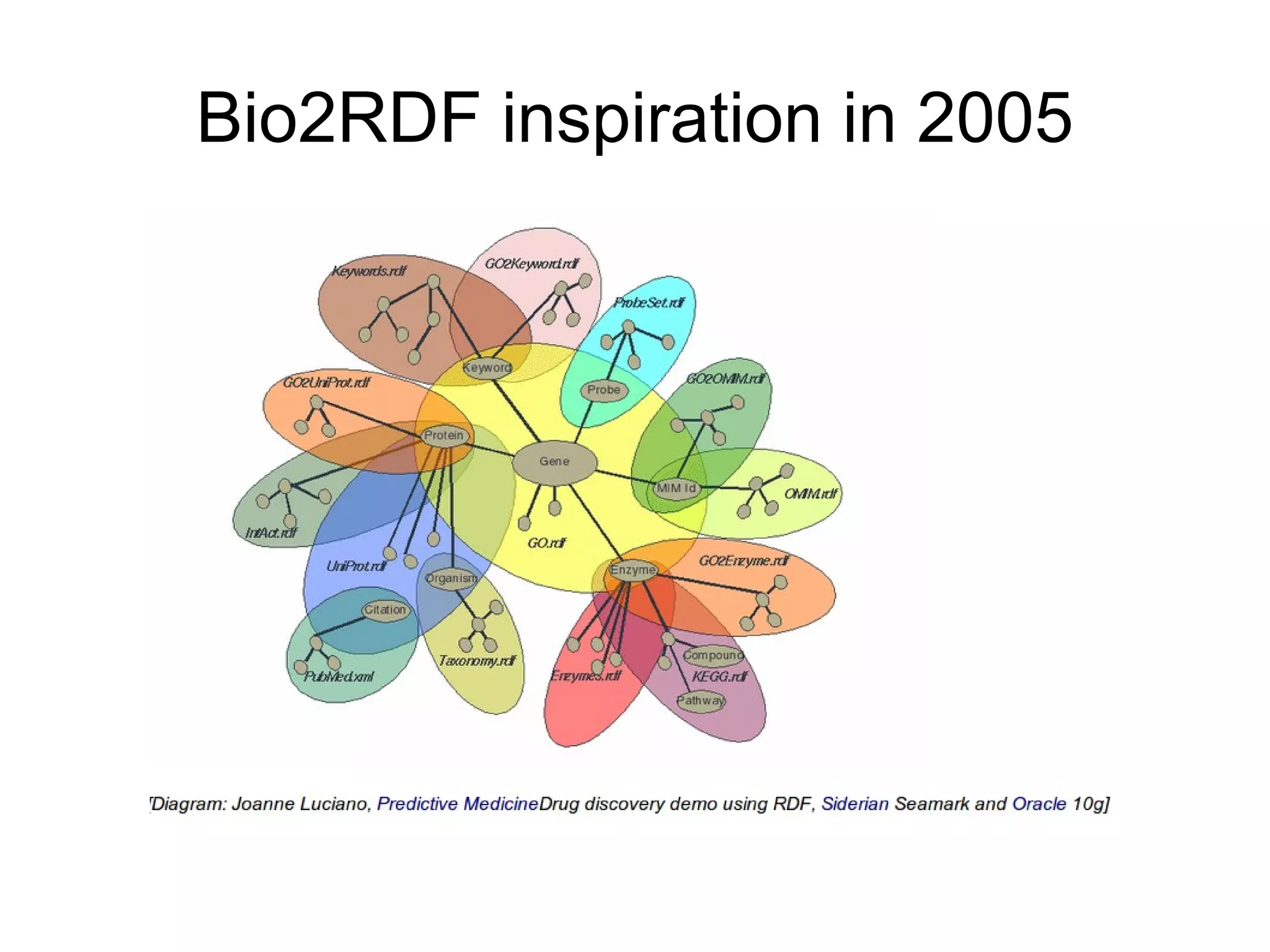
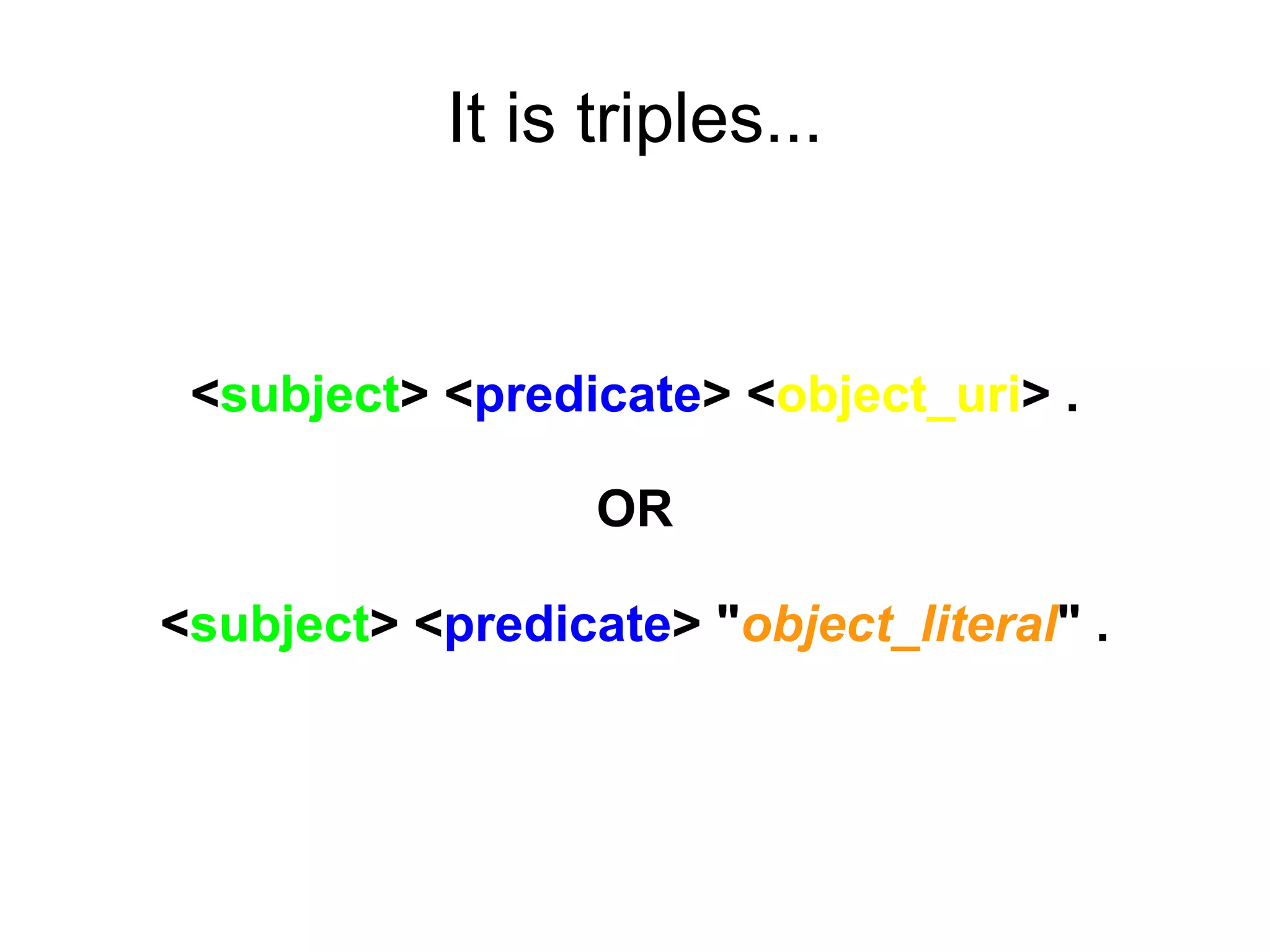
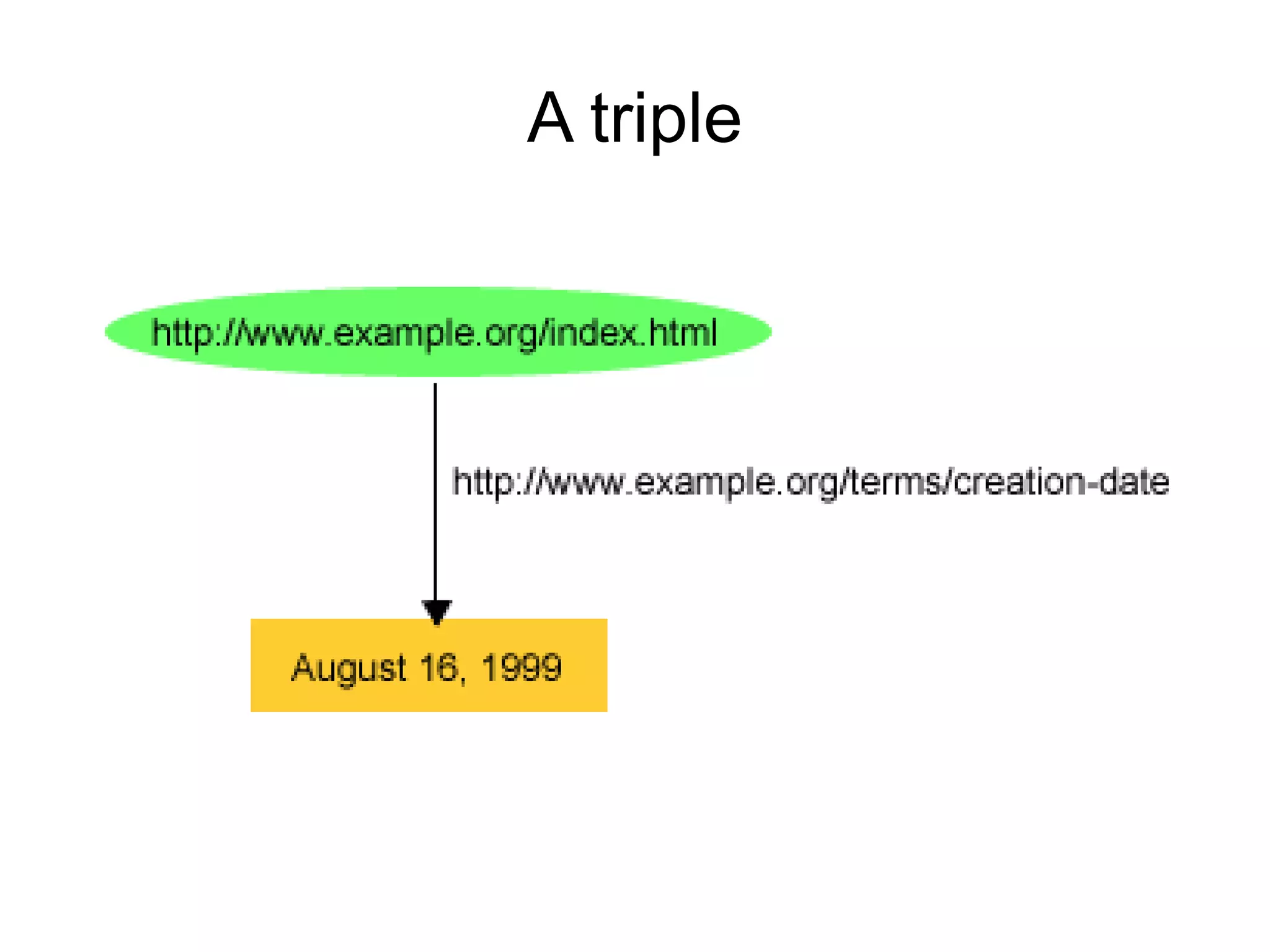
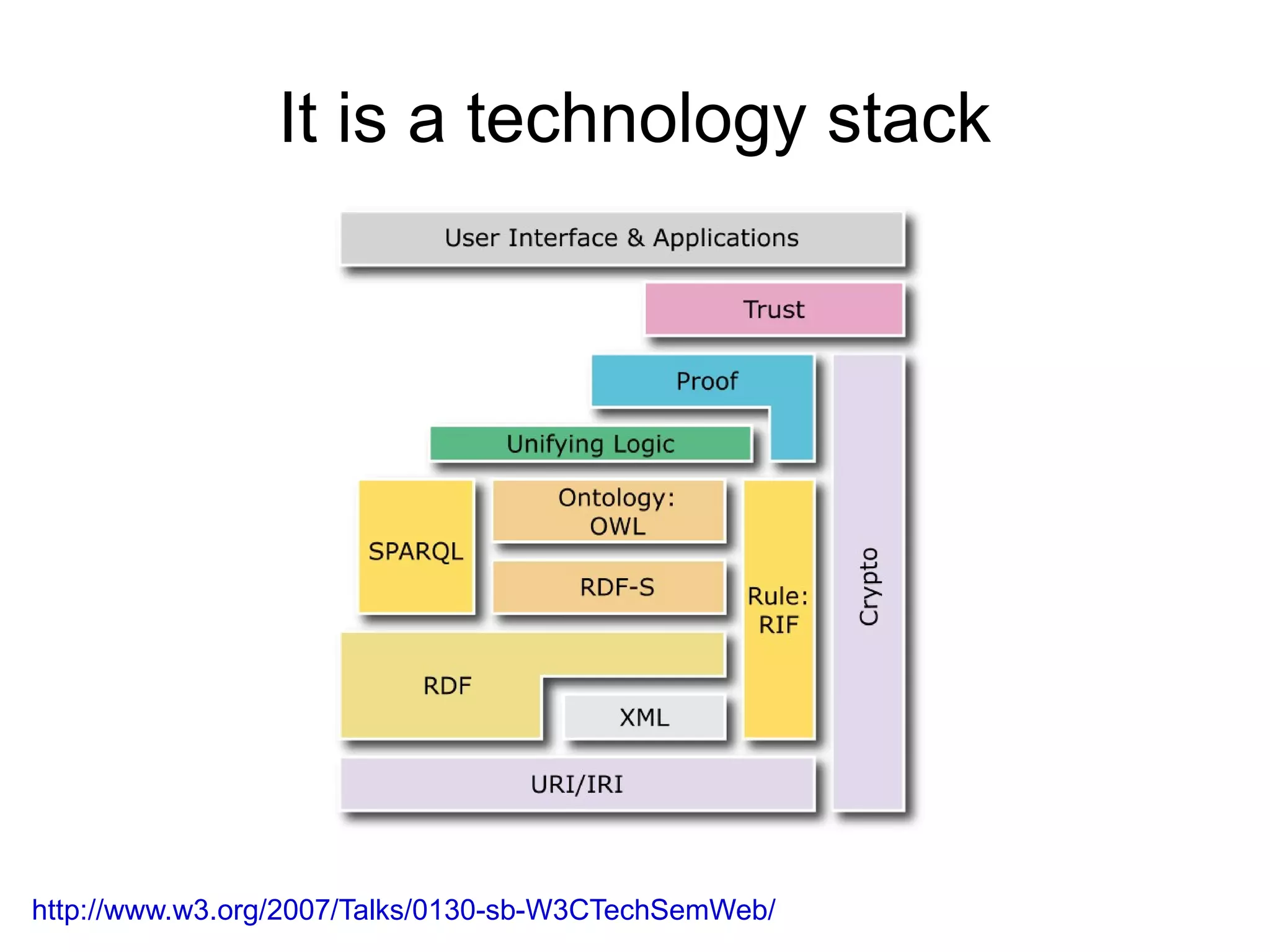
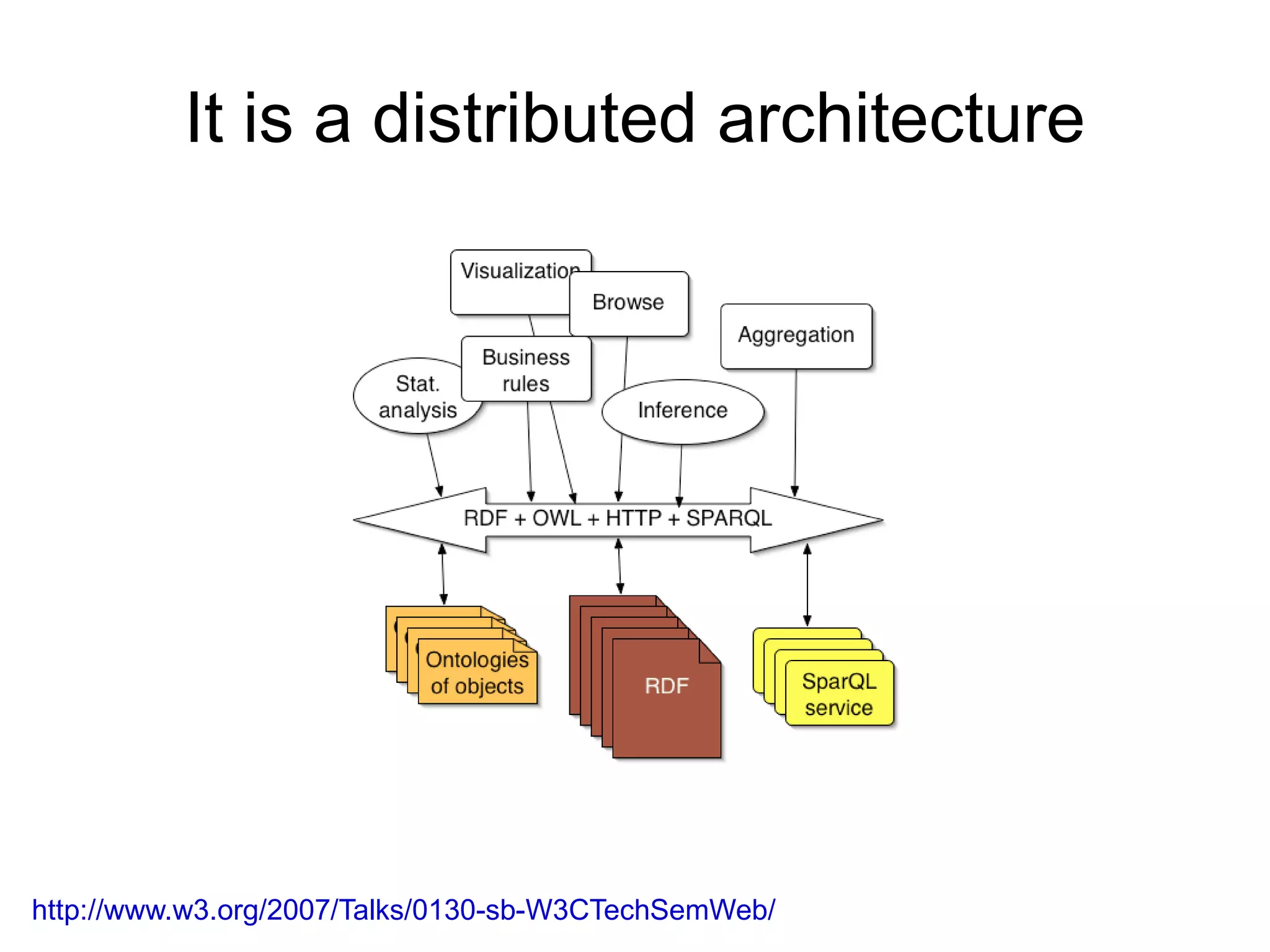
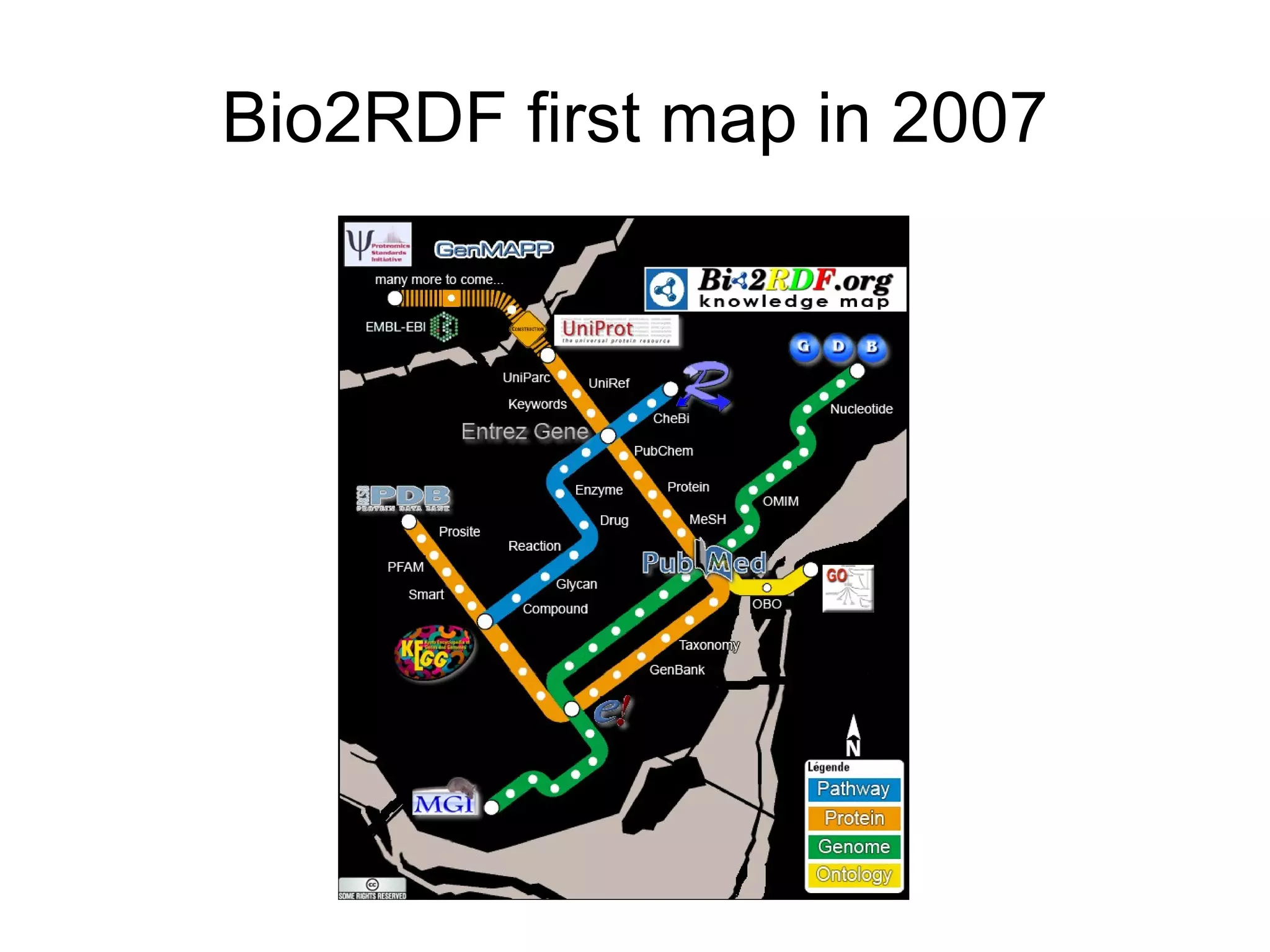
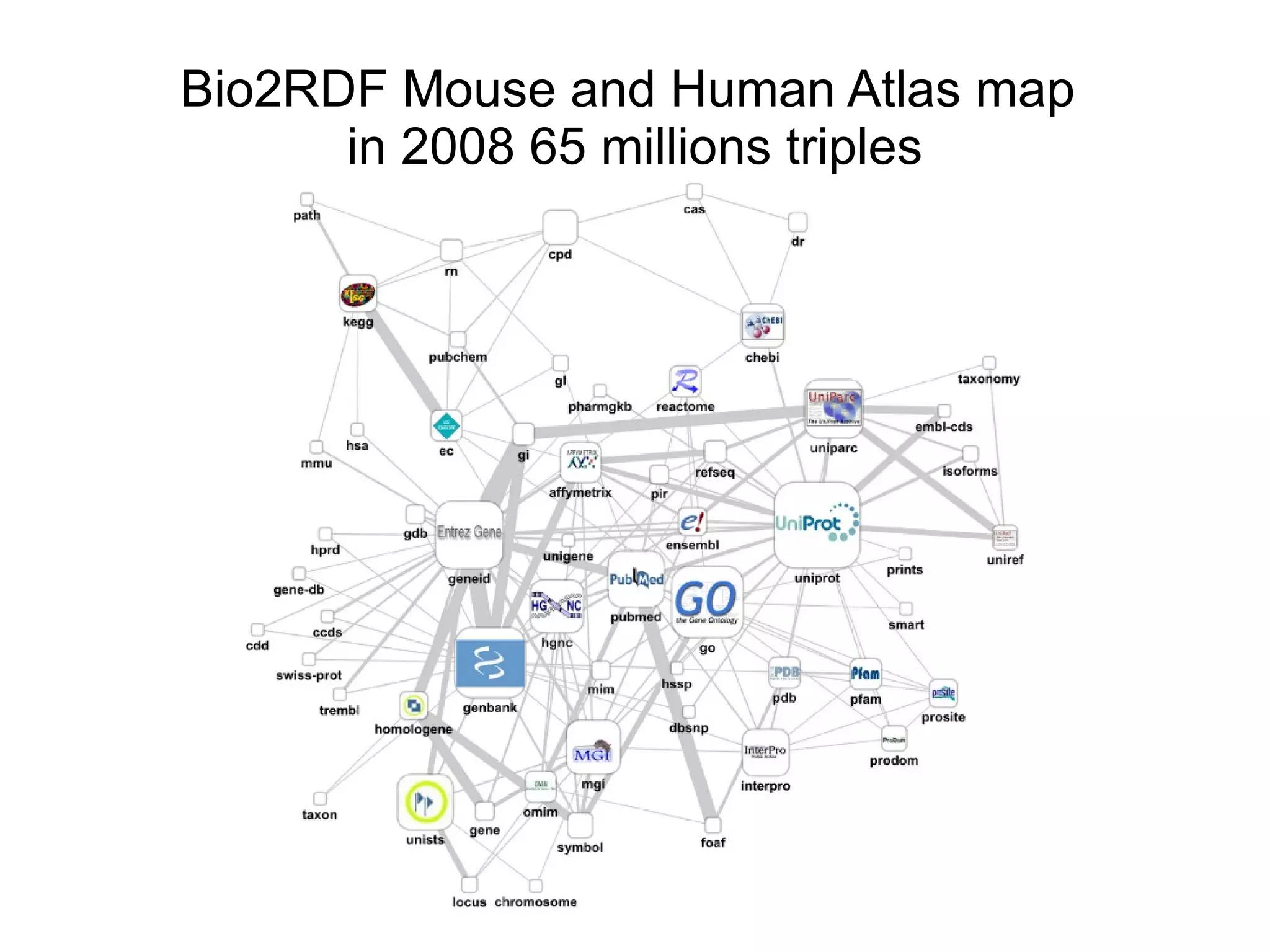
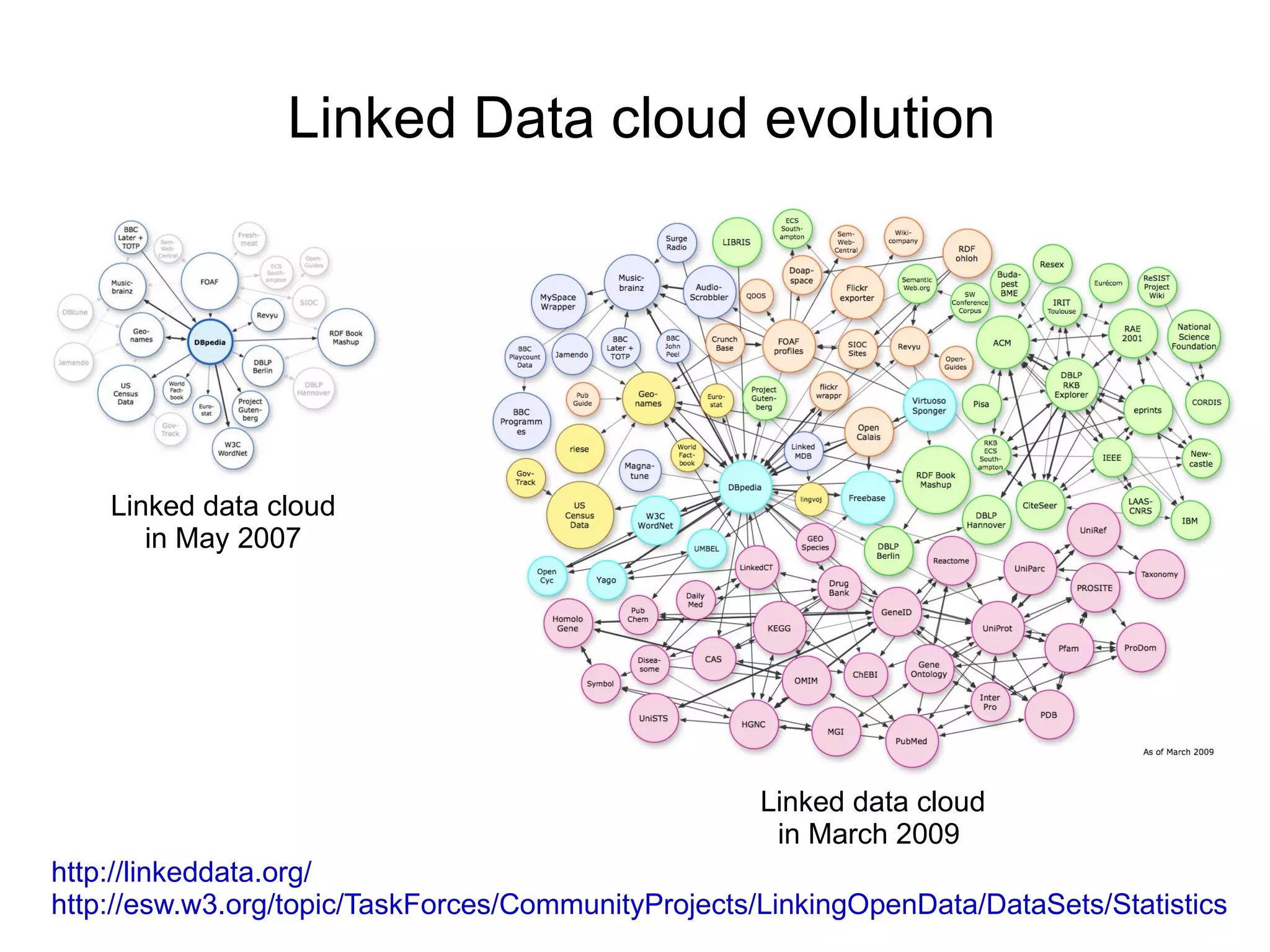
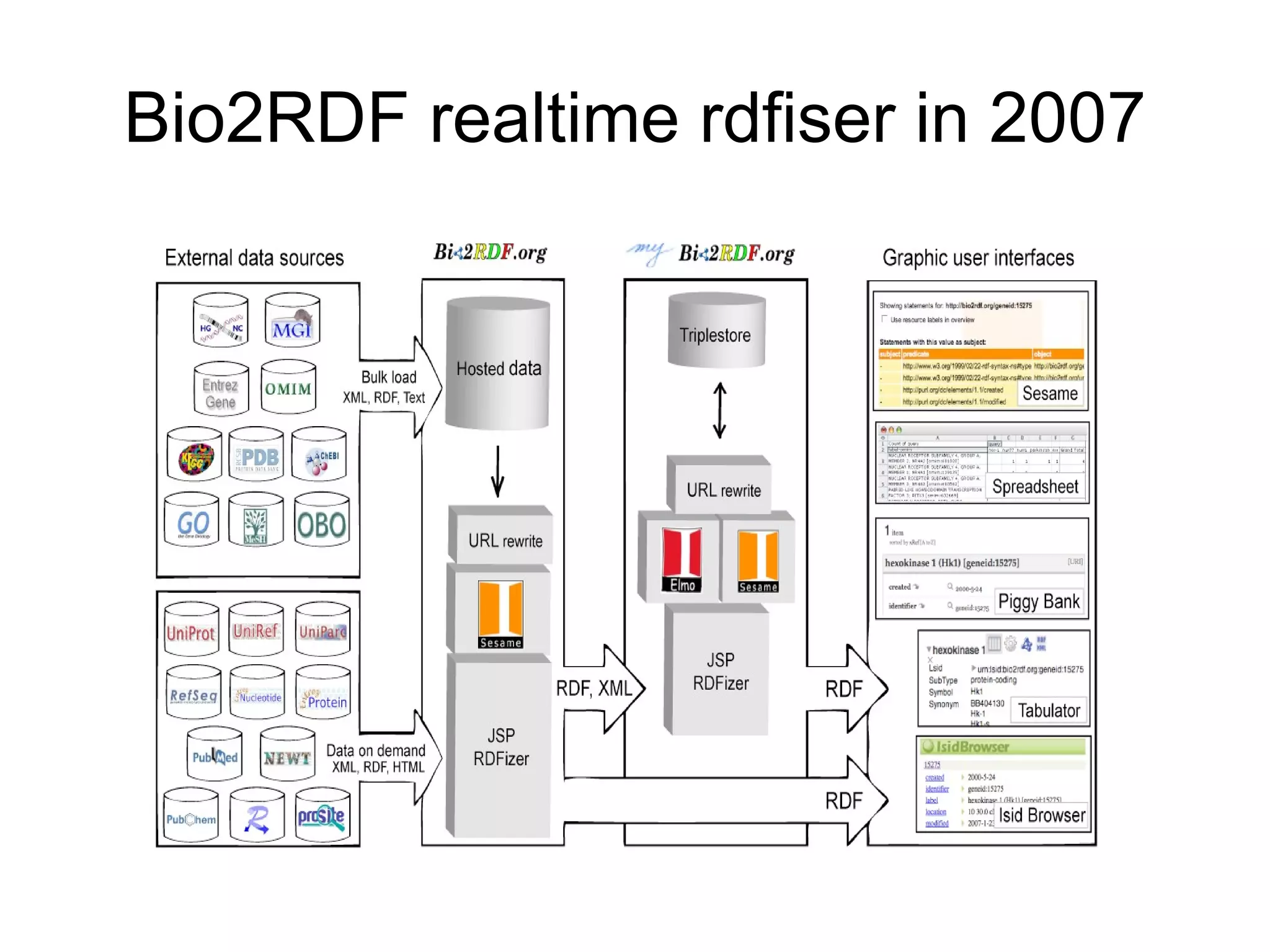
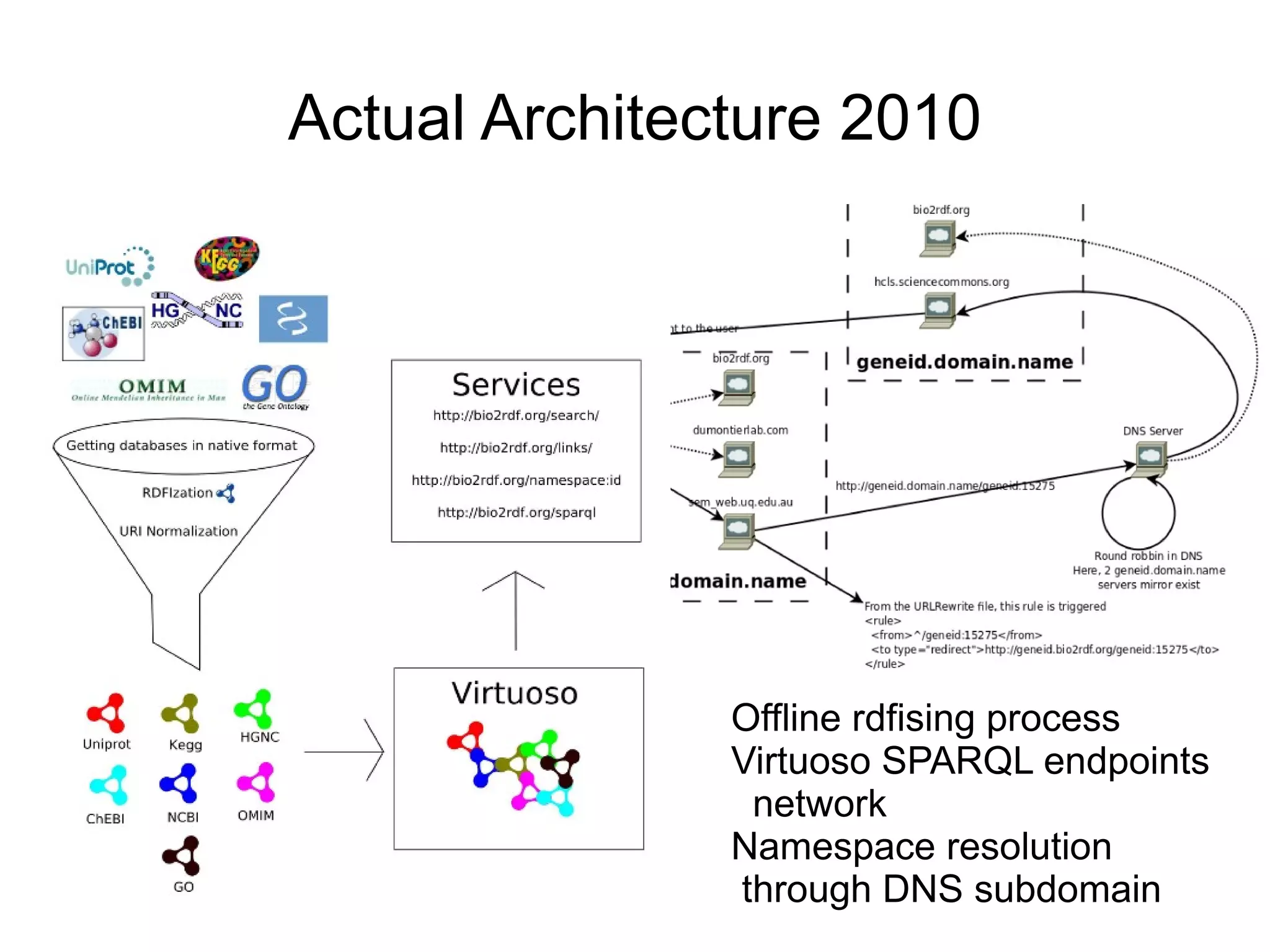
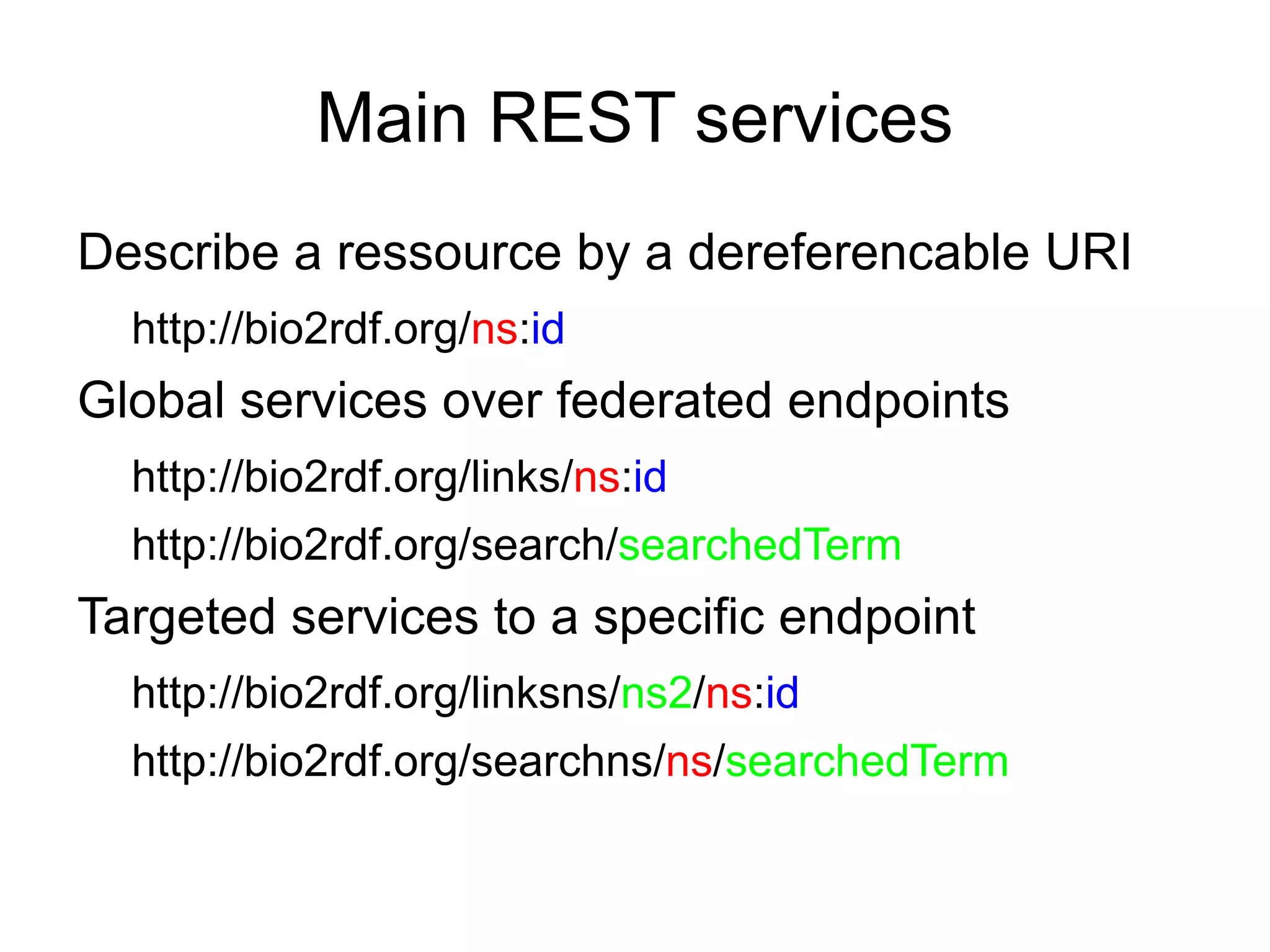
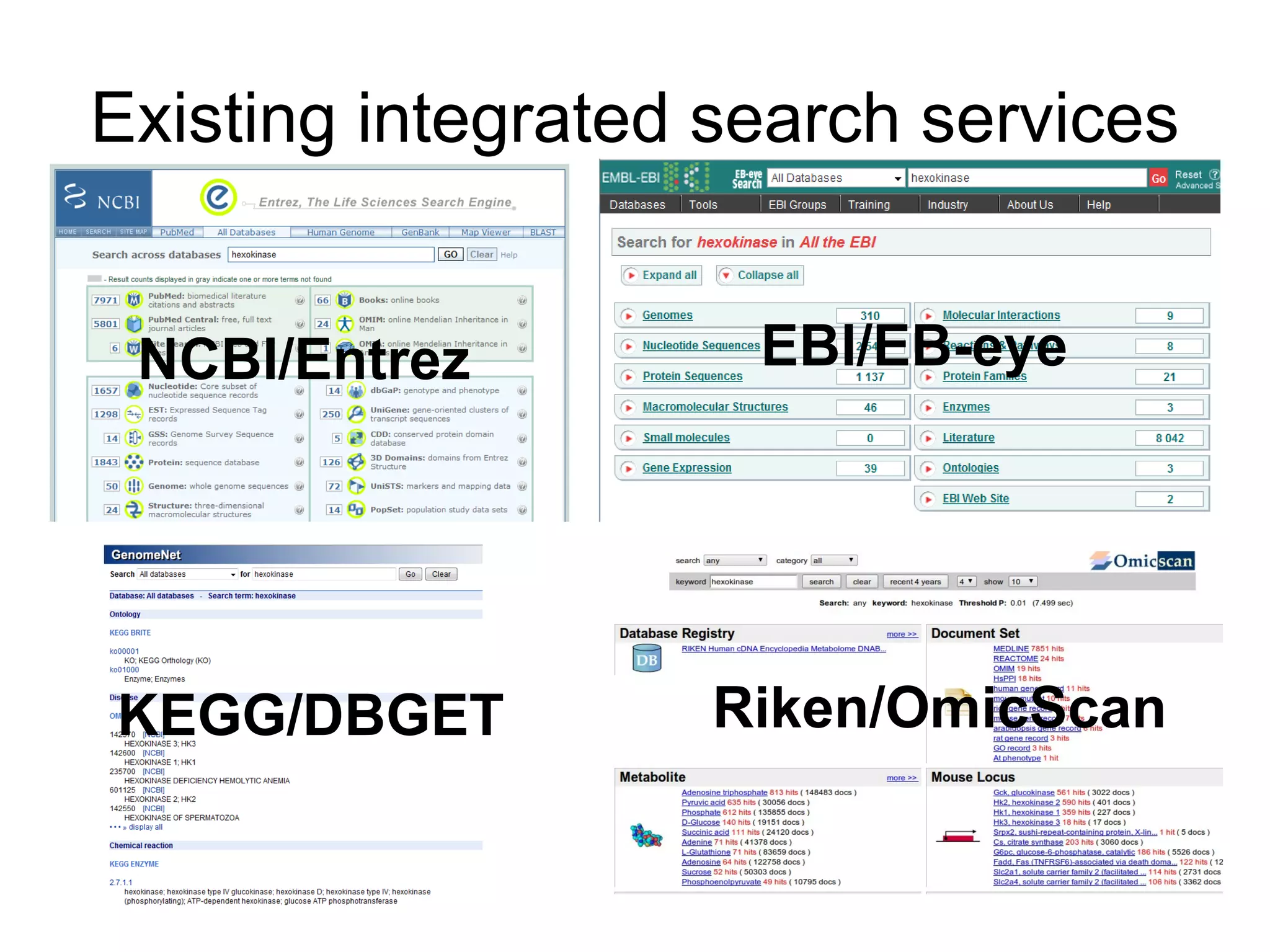
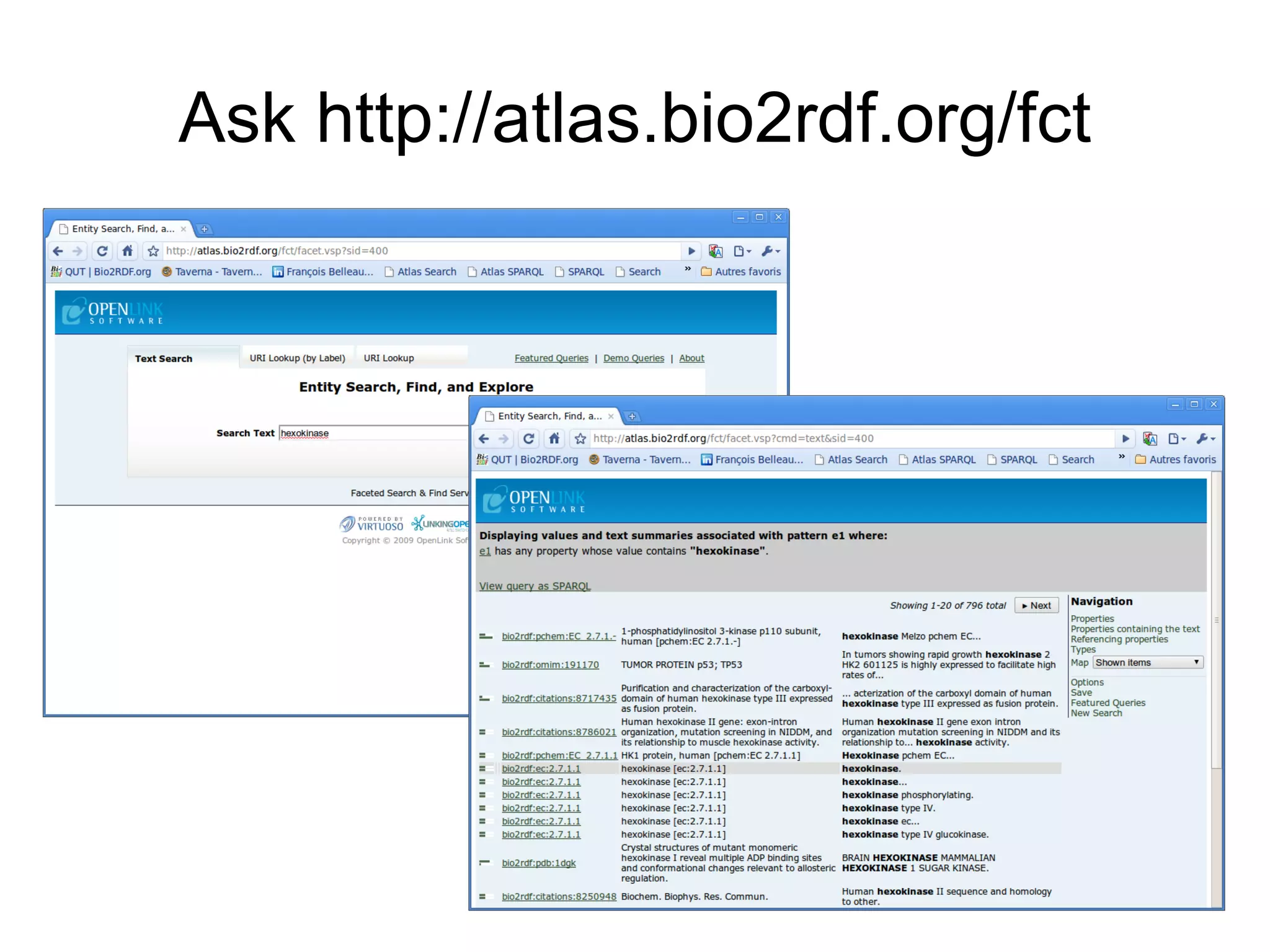
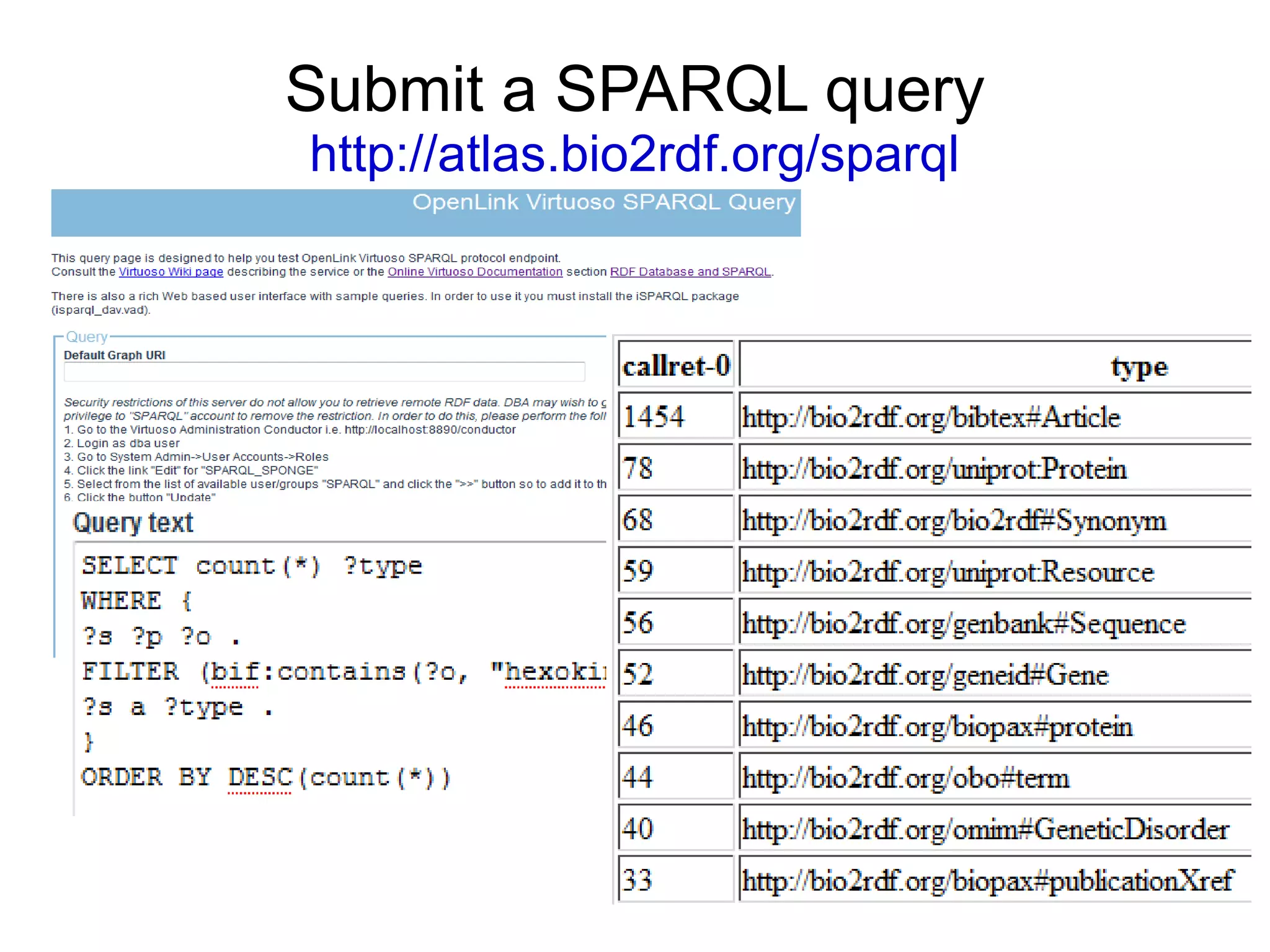
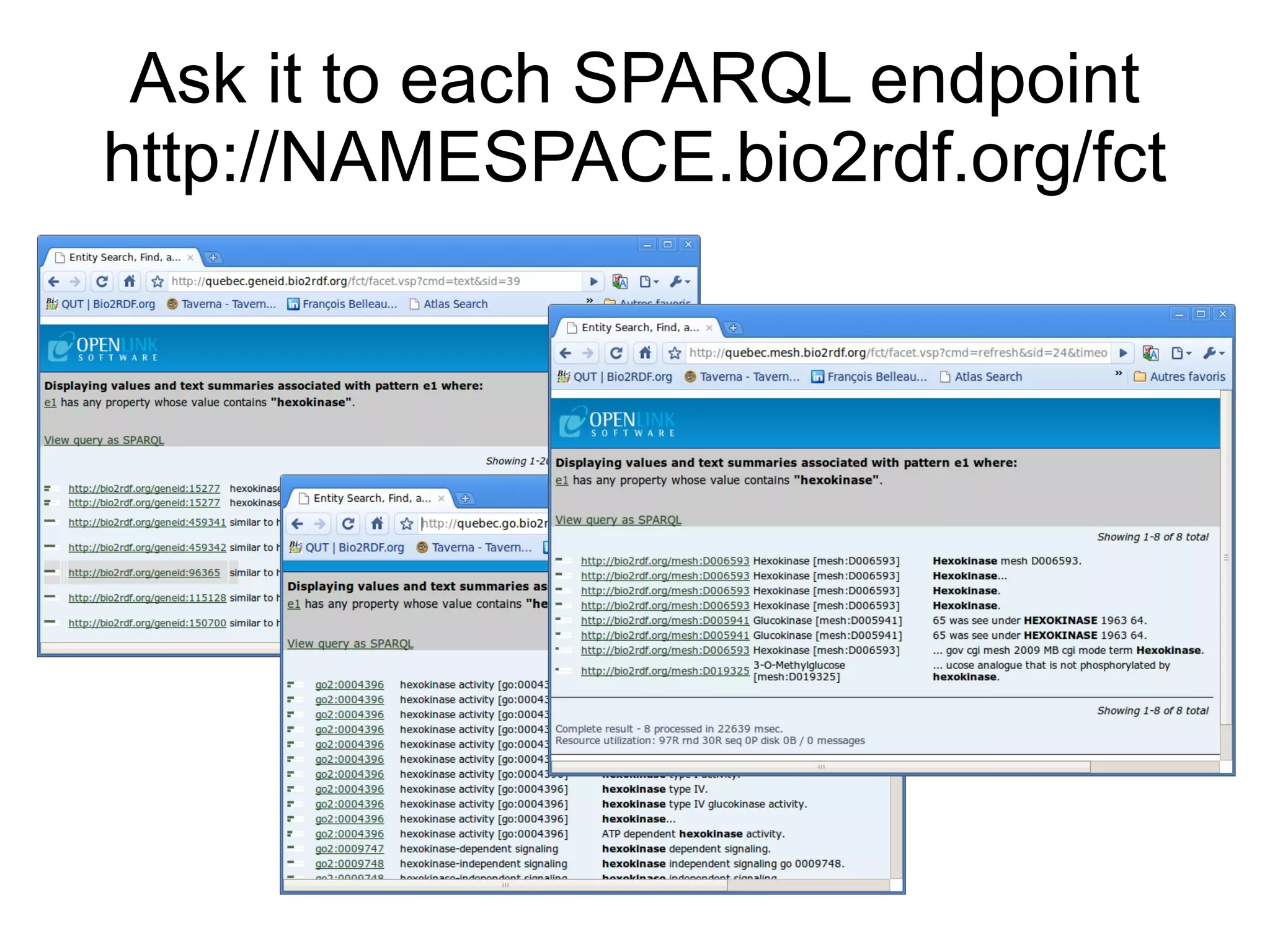
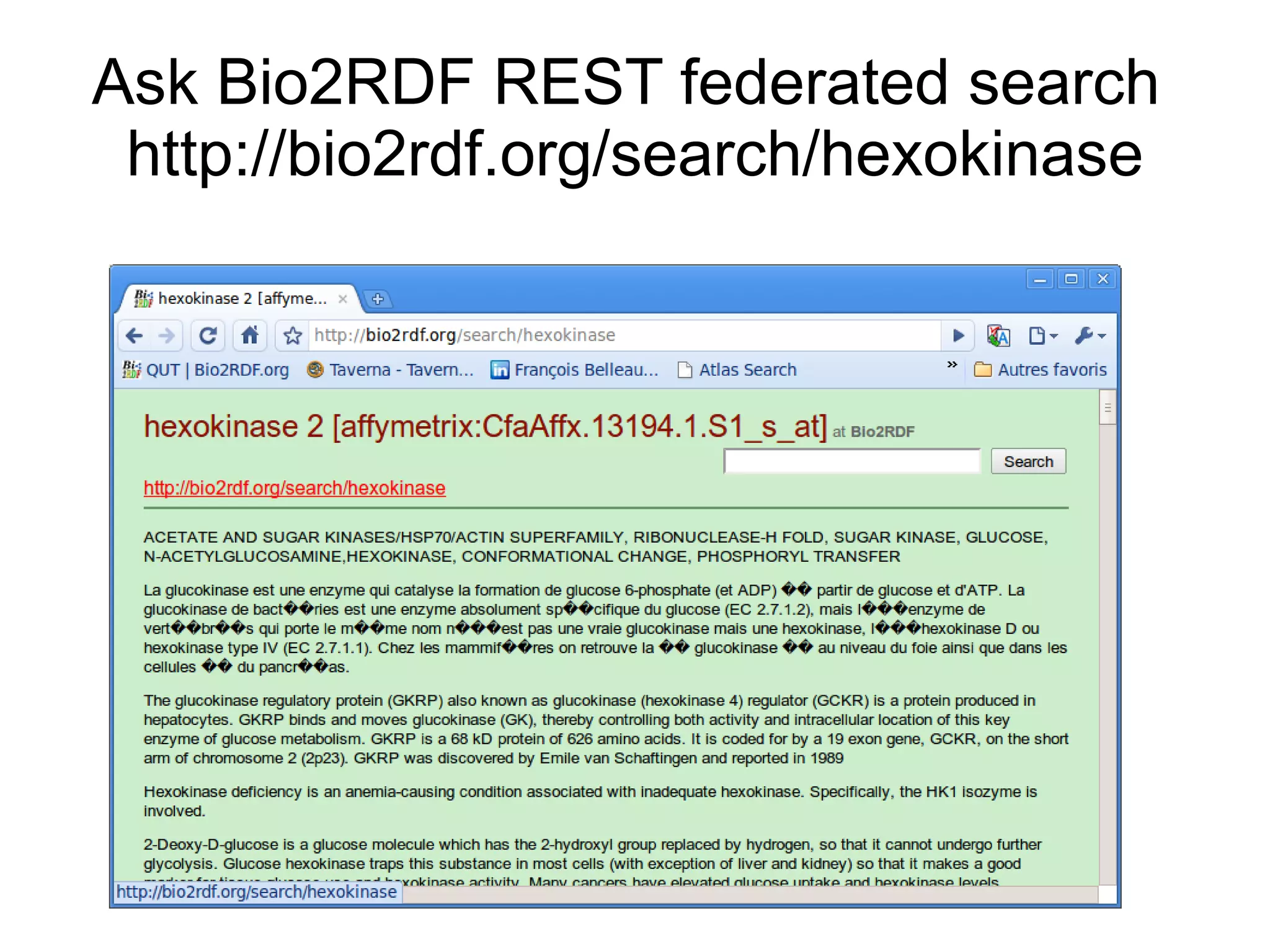
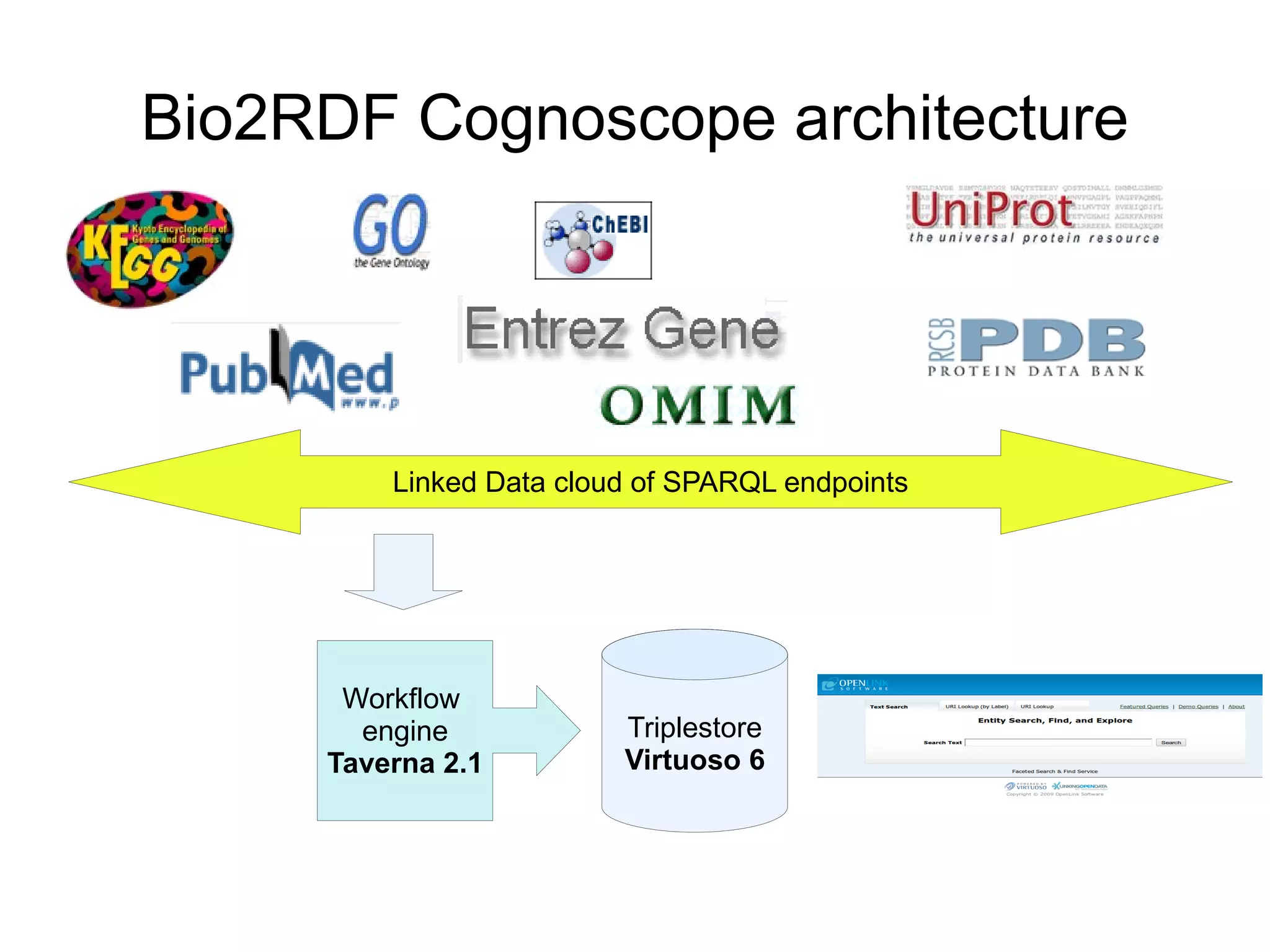
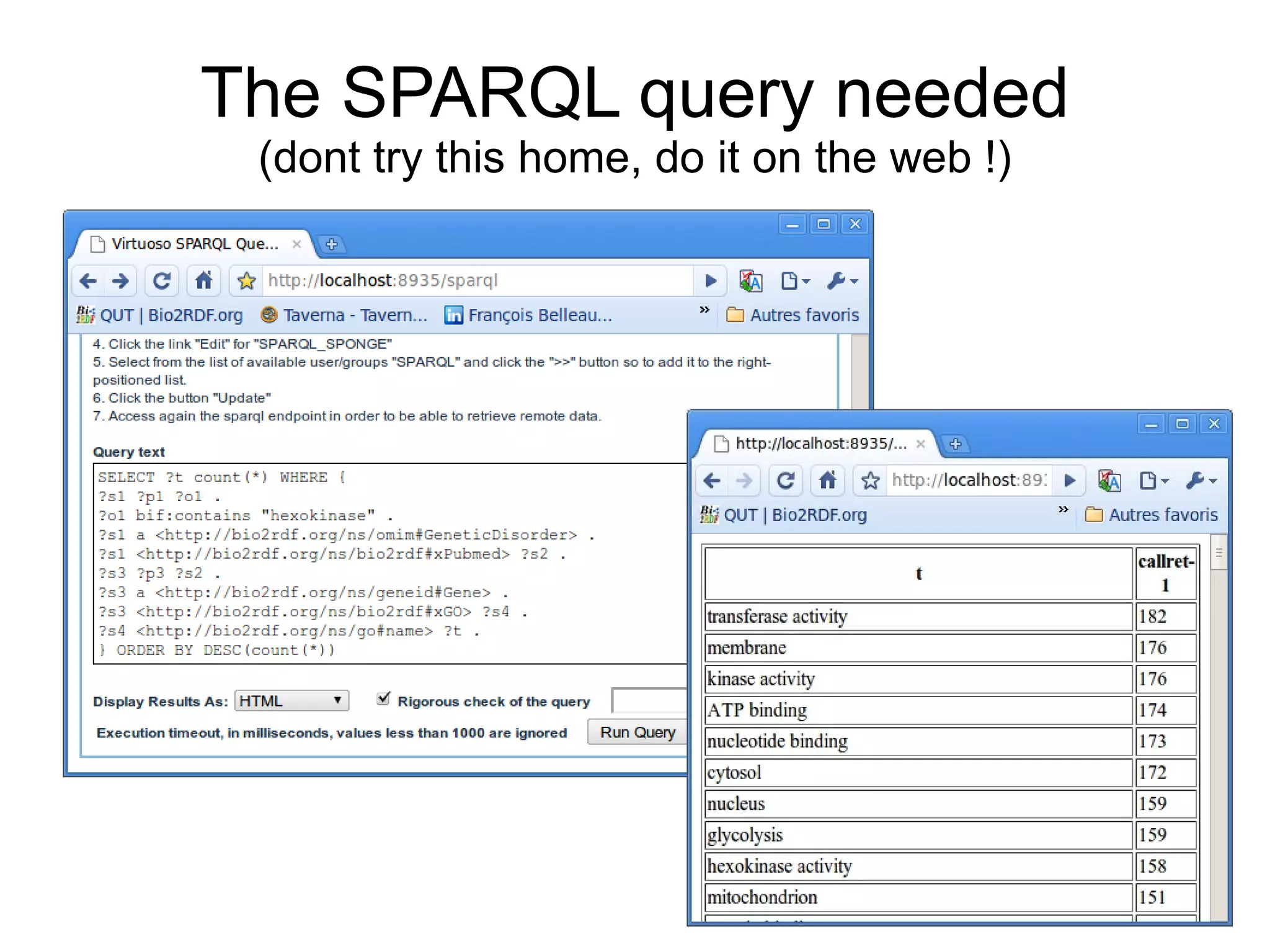
The document discusses Bio2RDF, a solution for data integration in bioinformatics using RDF, OWL, and SPARQL technologies, which aims to convert various public bioinformatics databases into RDF format. It introduces the concept of 'Cognoscope,' a tool designed to query a network of SPARQL endpoints to extract knowledge about specific entities, like hexokinase, through advanced query workflows. The document emphasizes the importance of building a critical mass of ontologies in life sciences for rapid semantic web adoption and development.