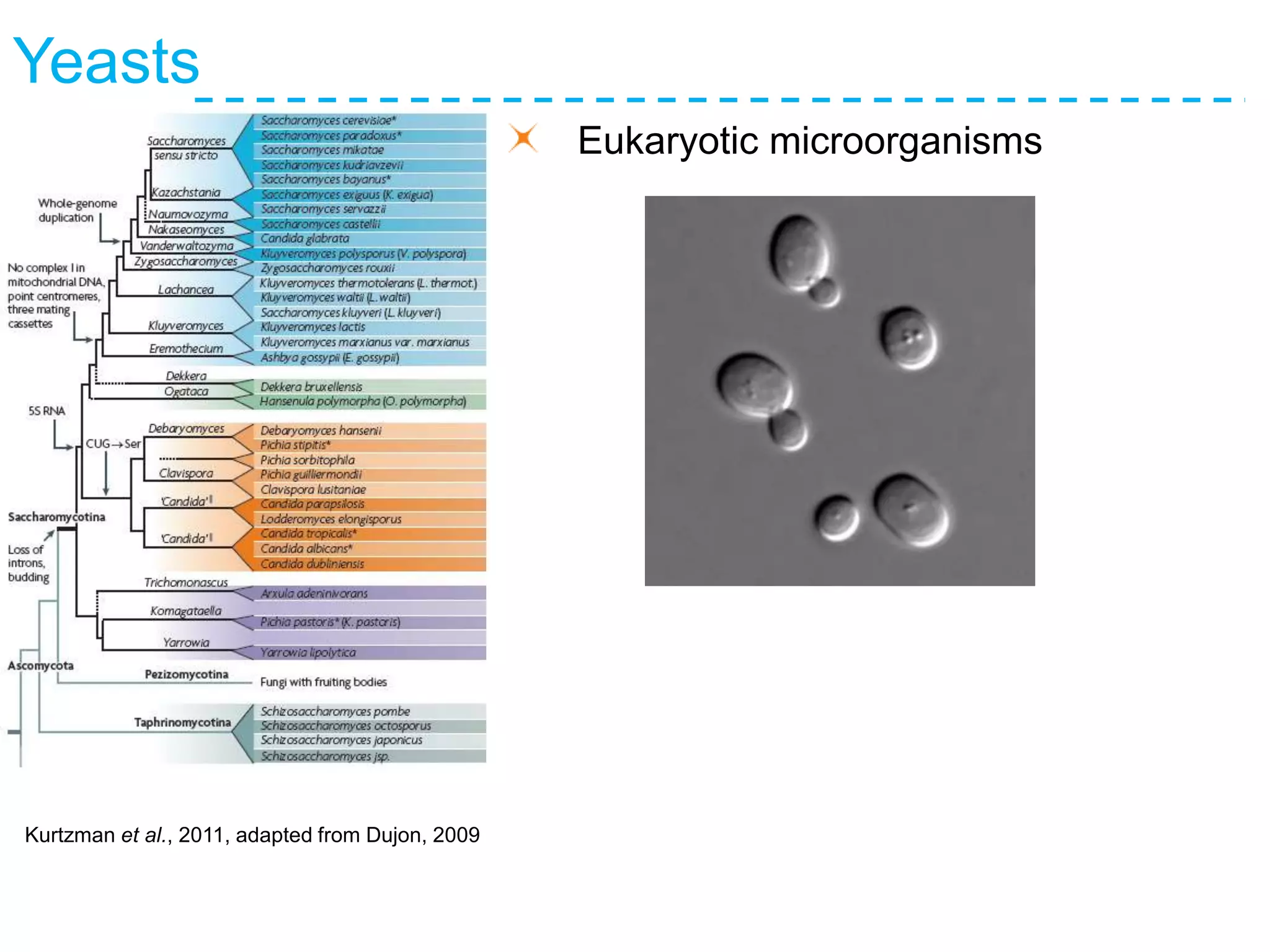
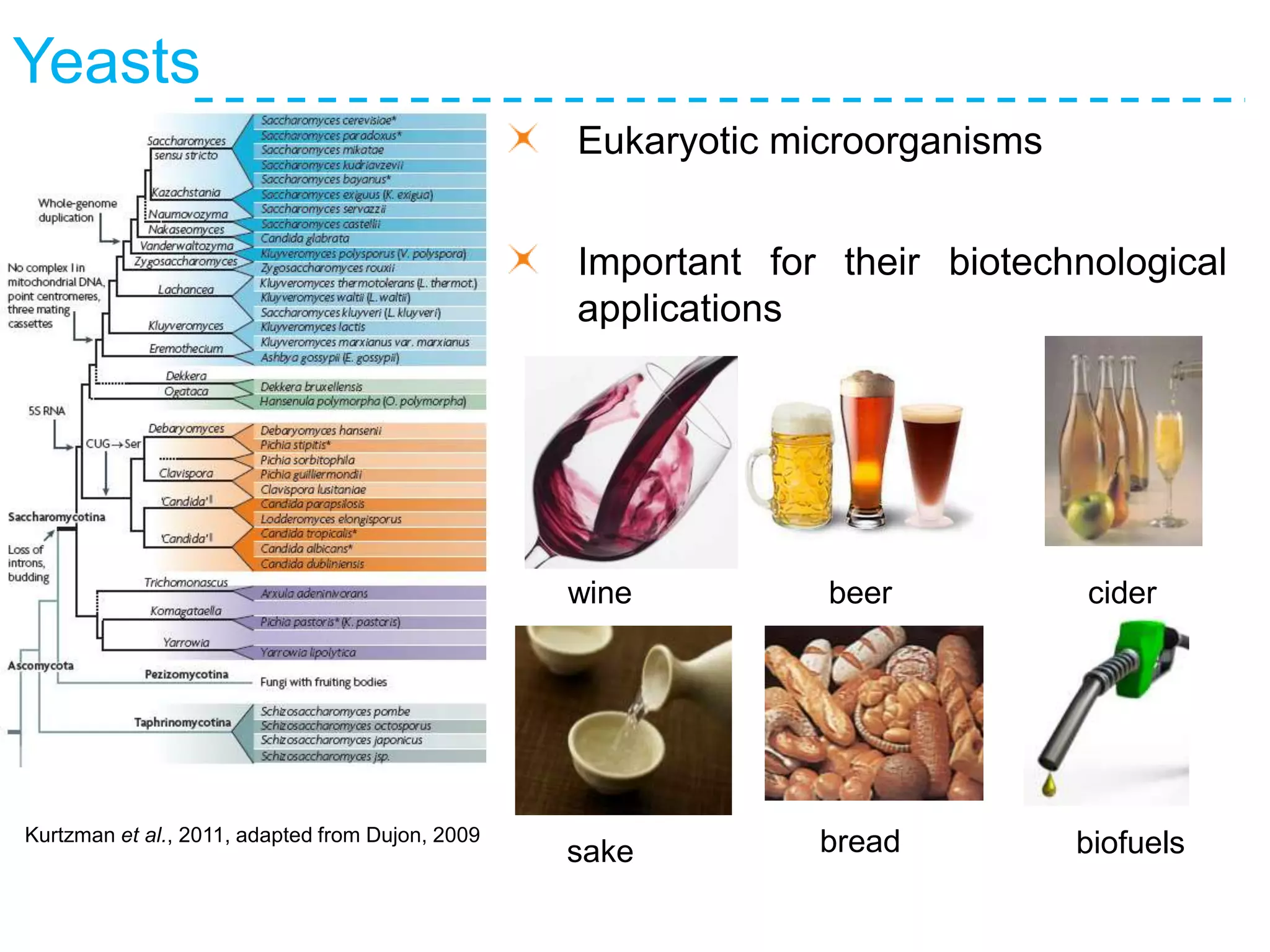
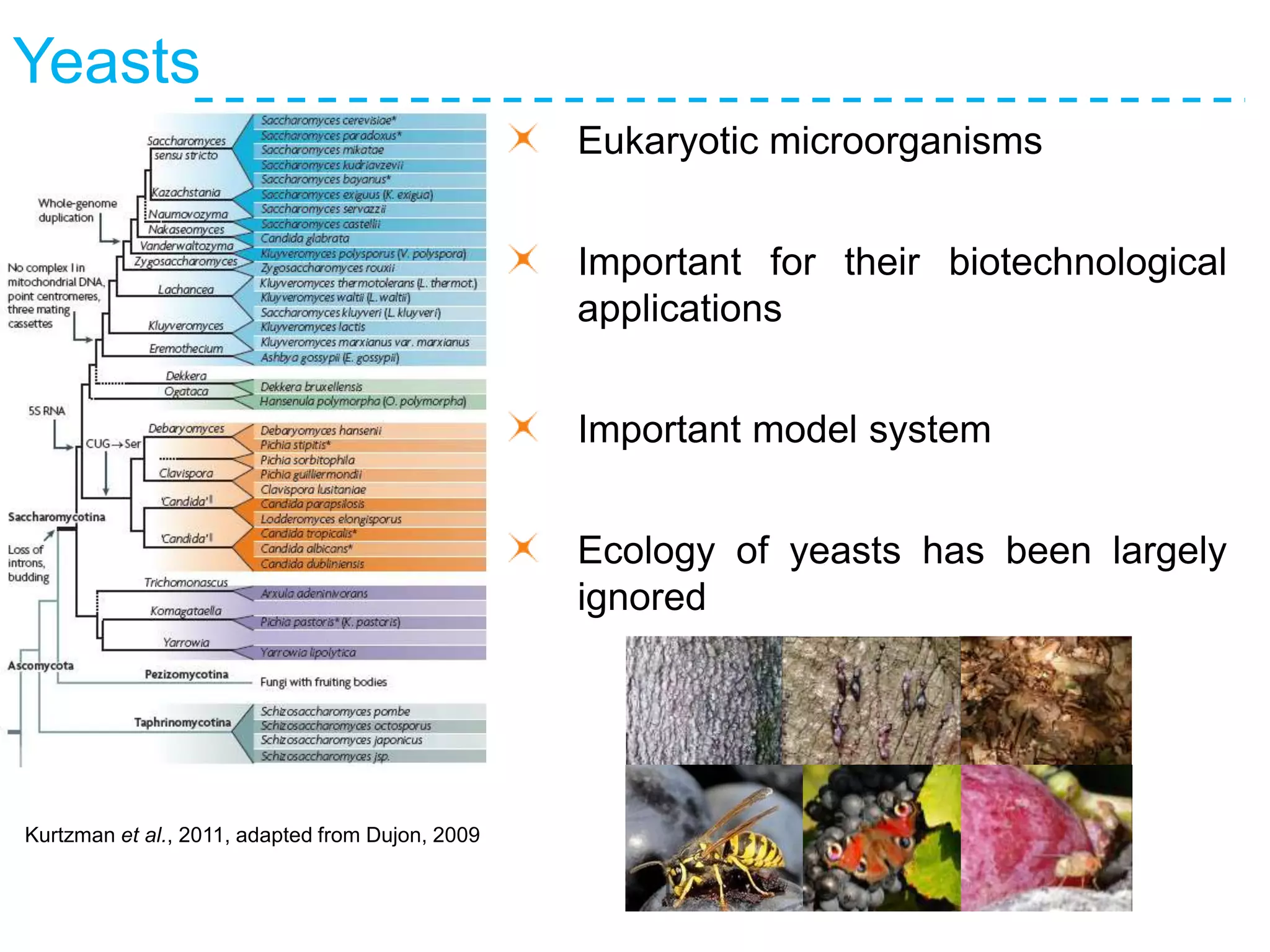
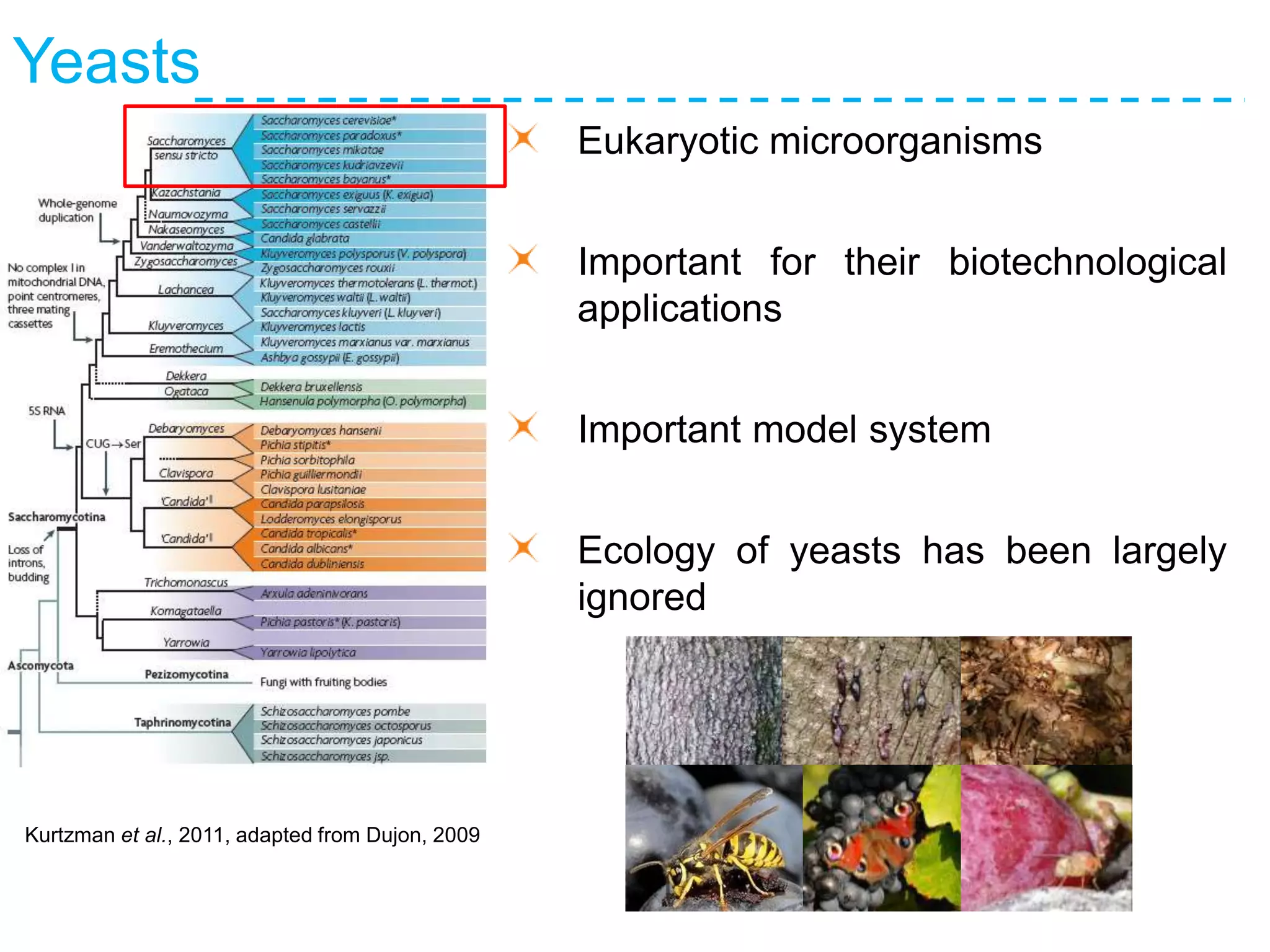
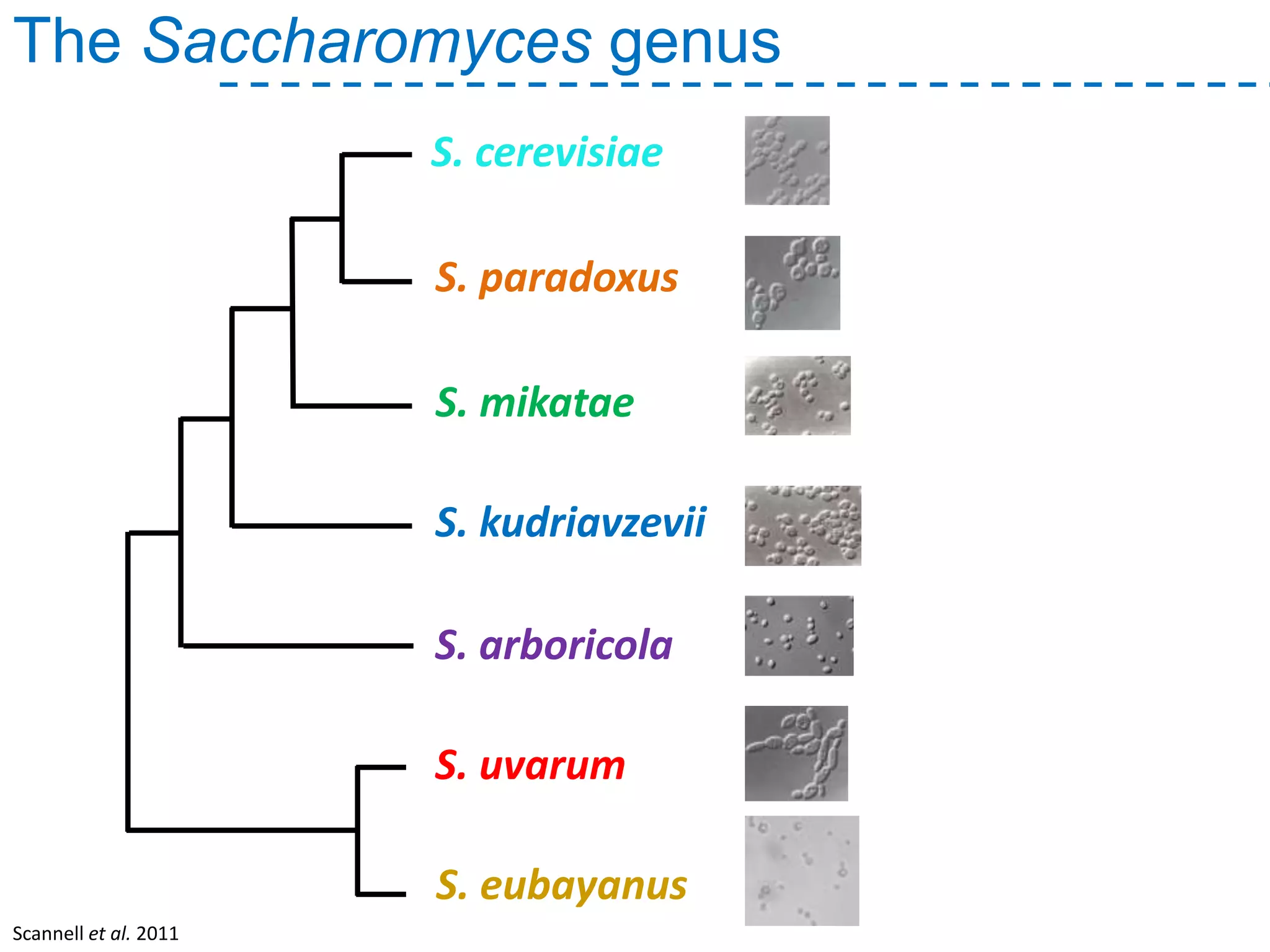
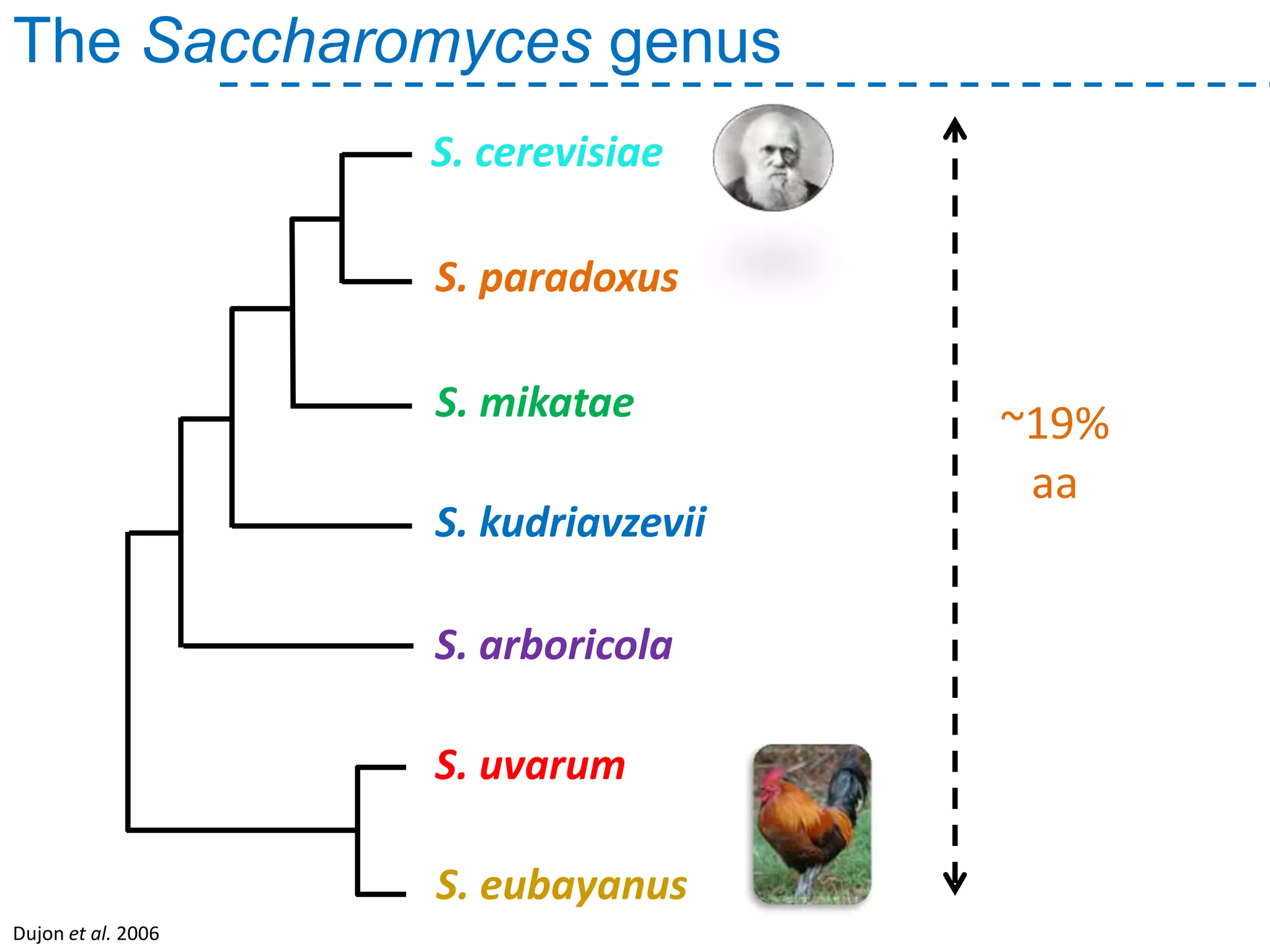
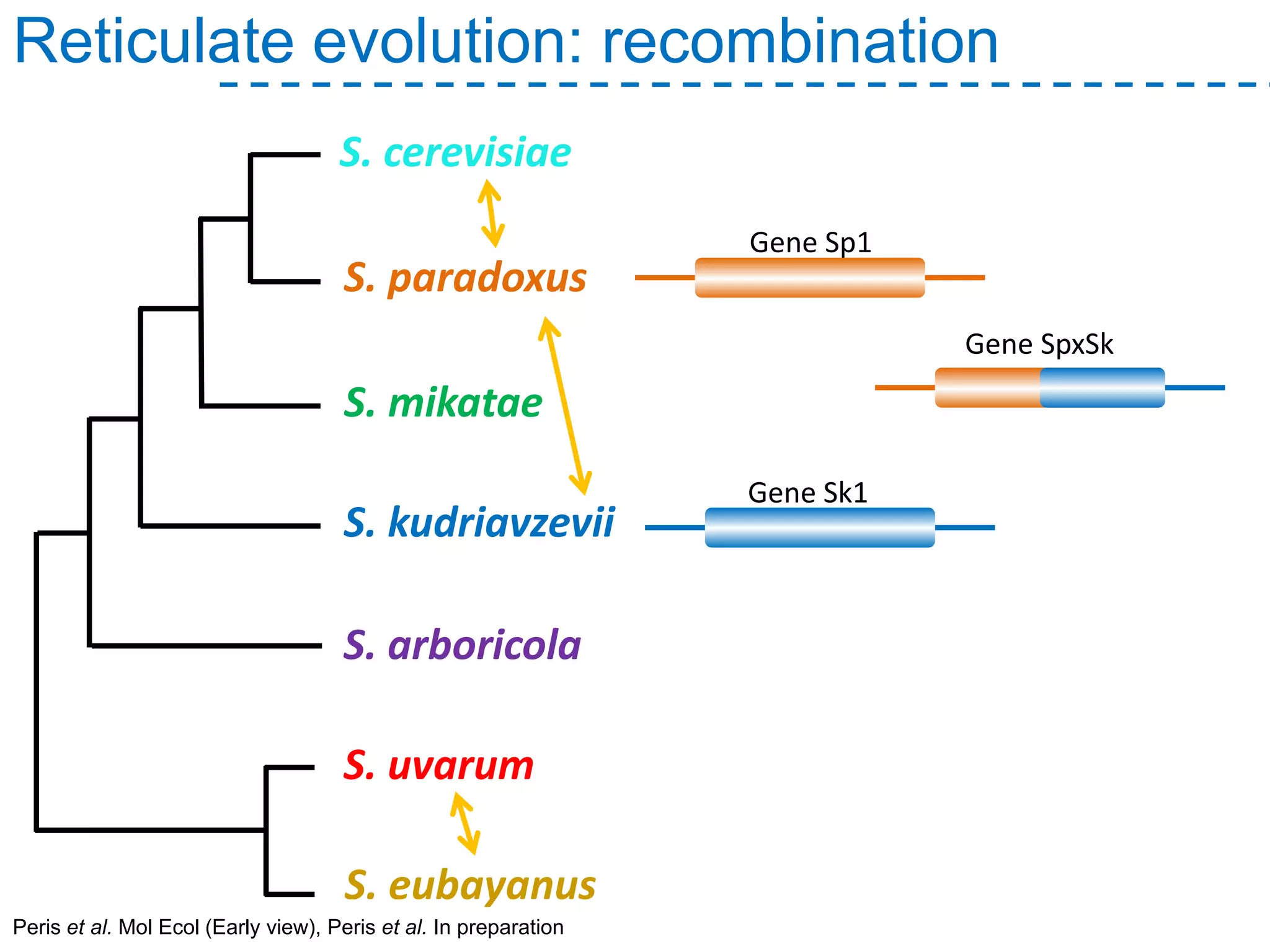
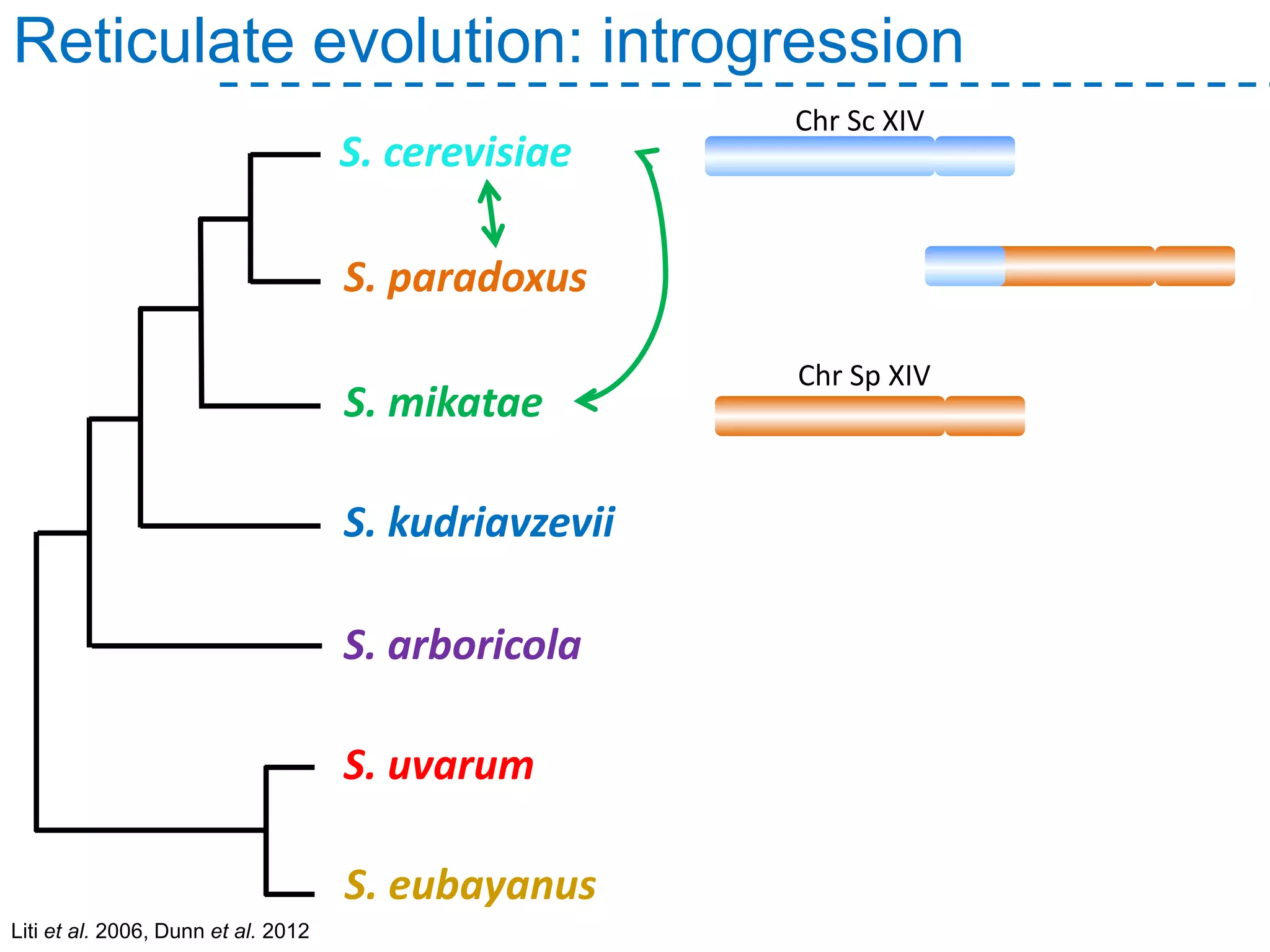
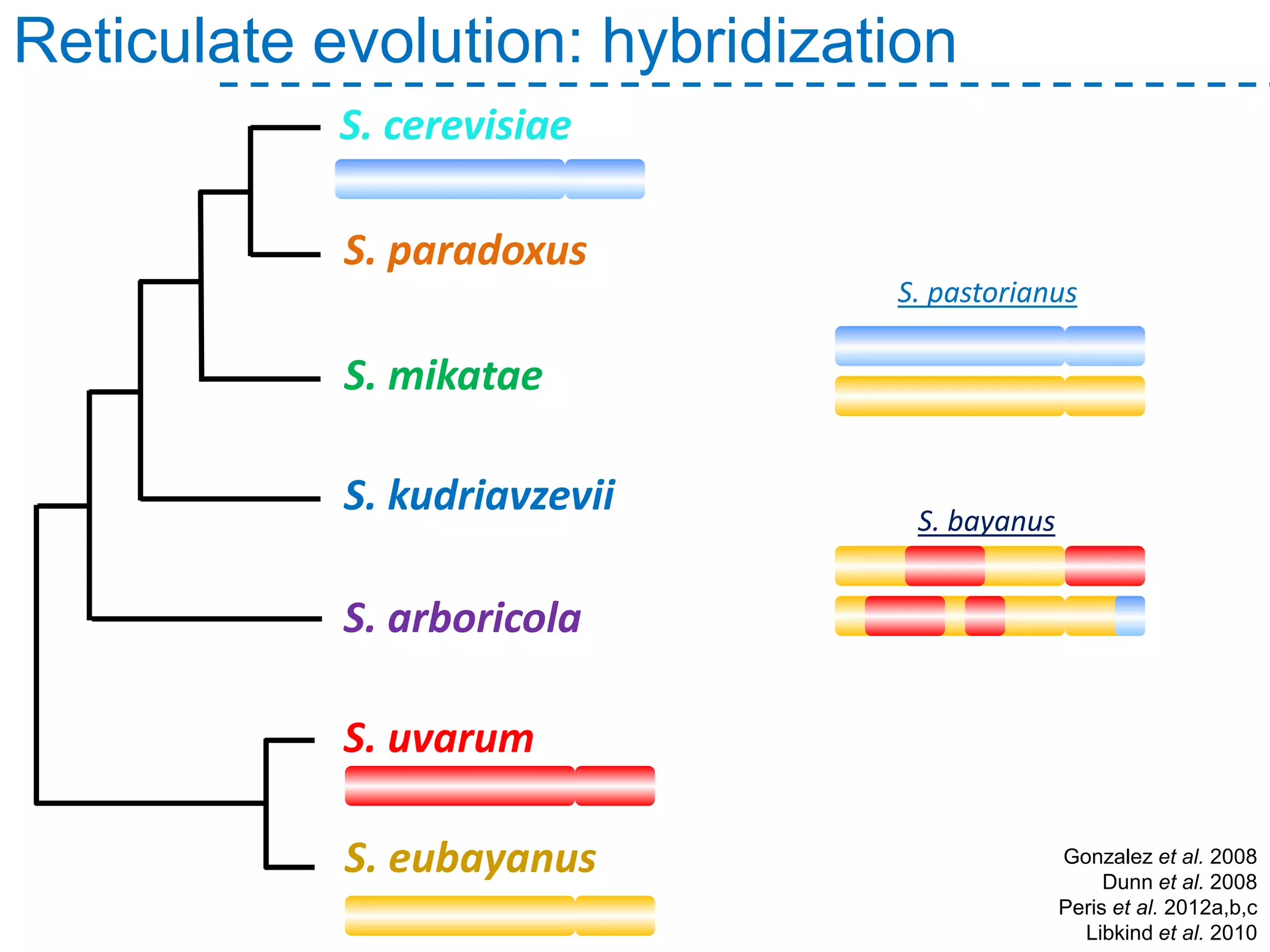
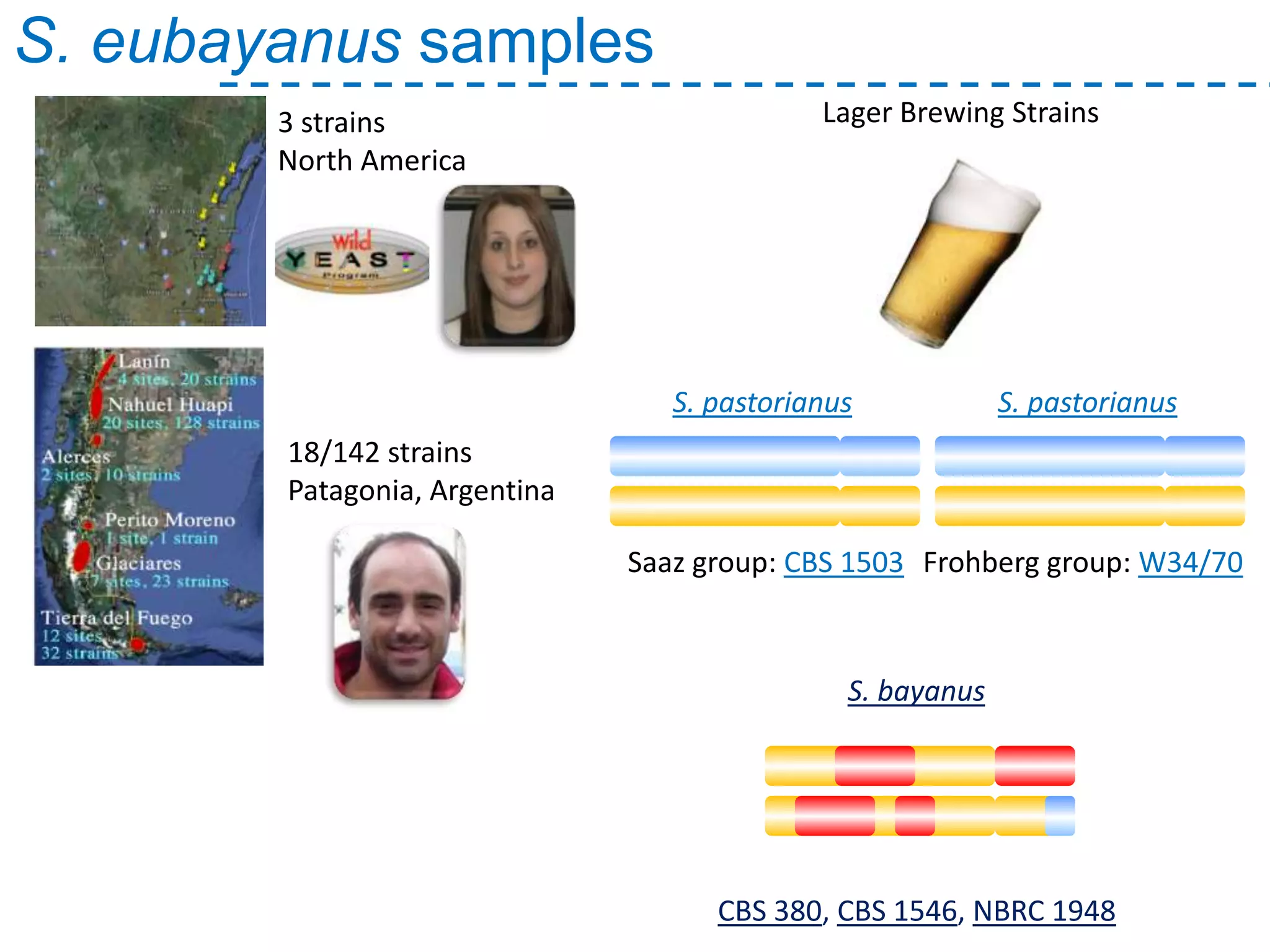
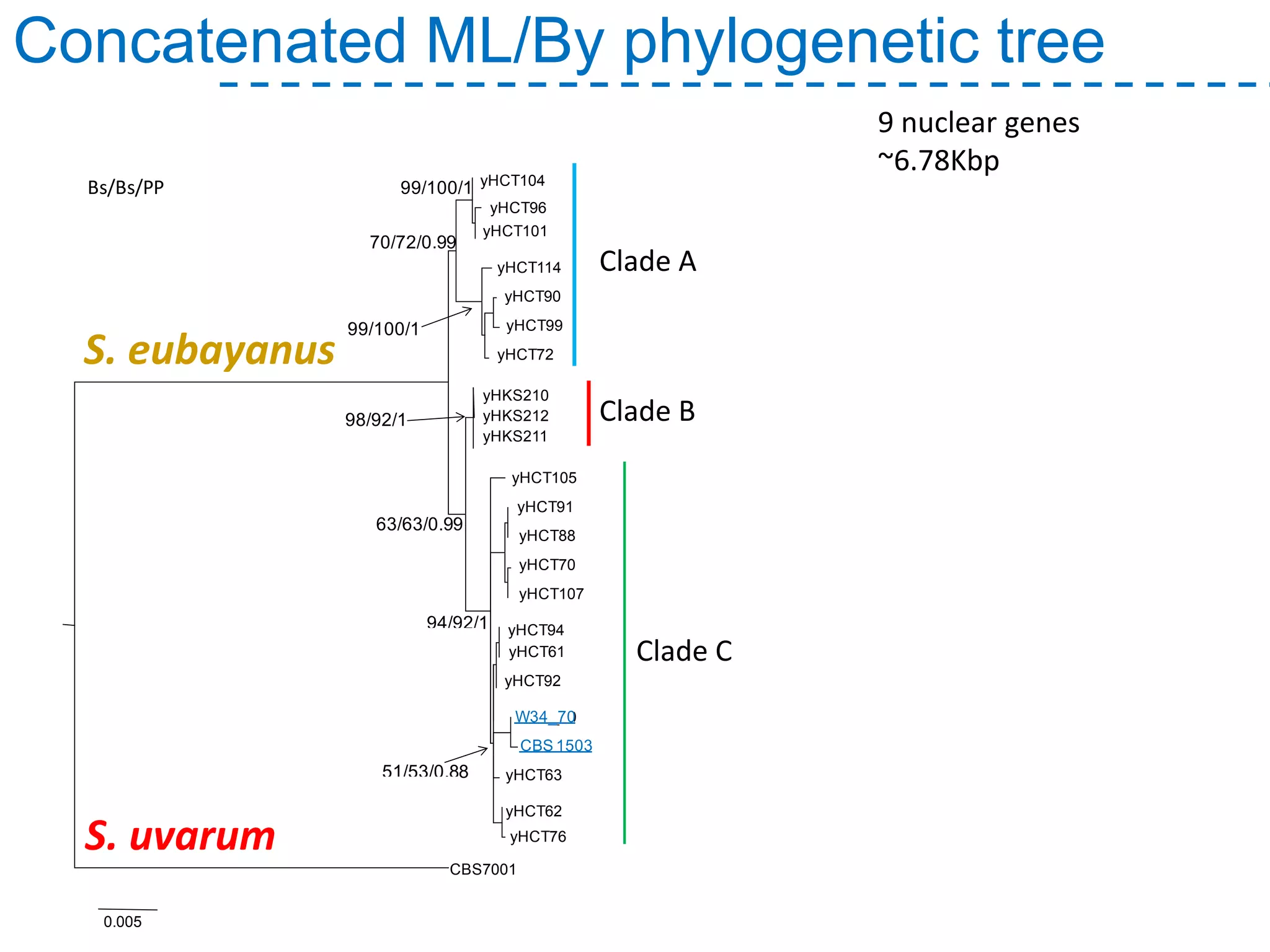
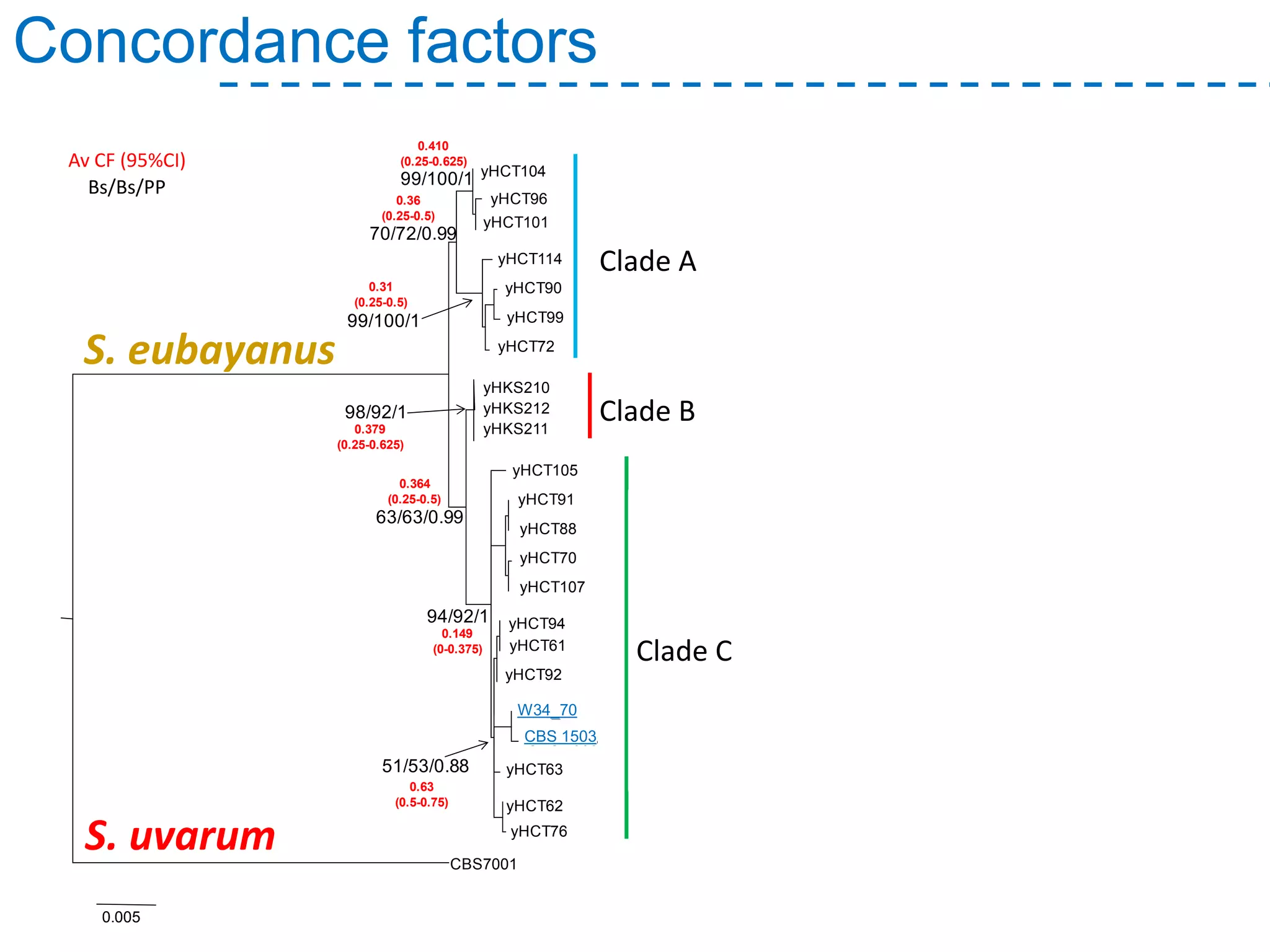
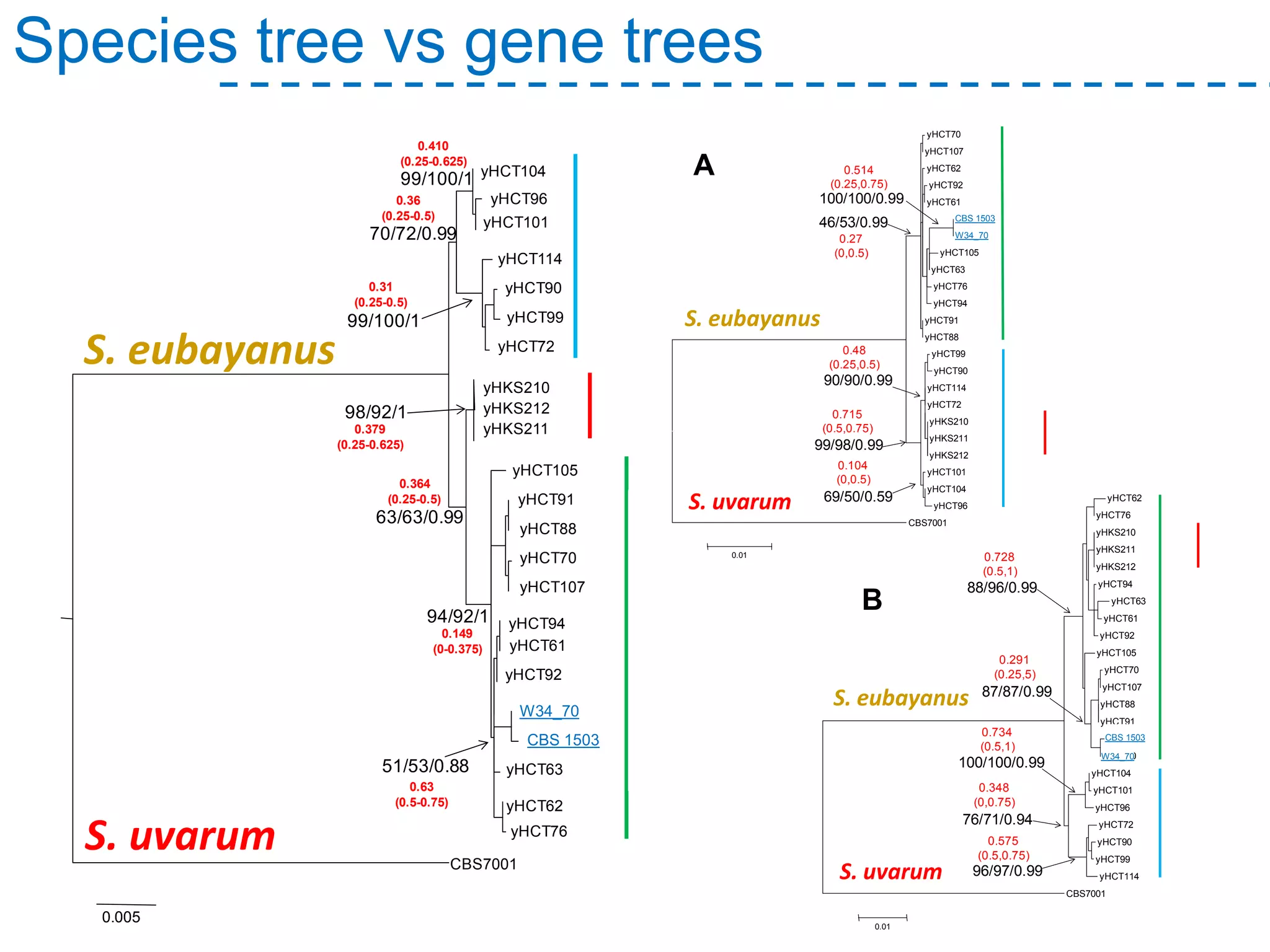
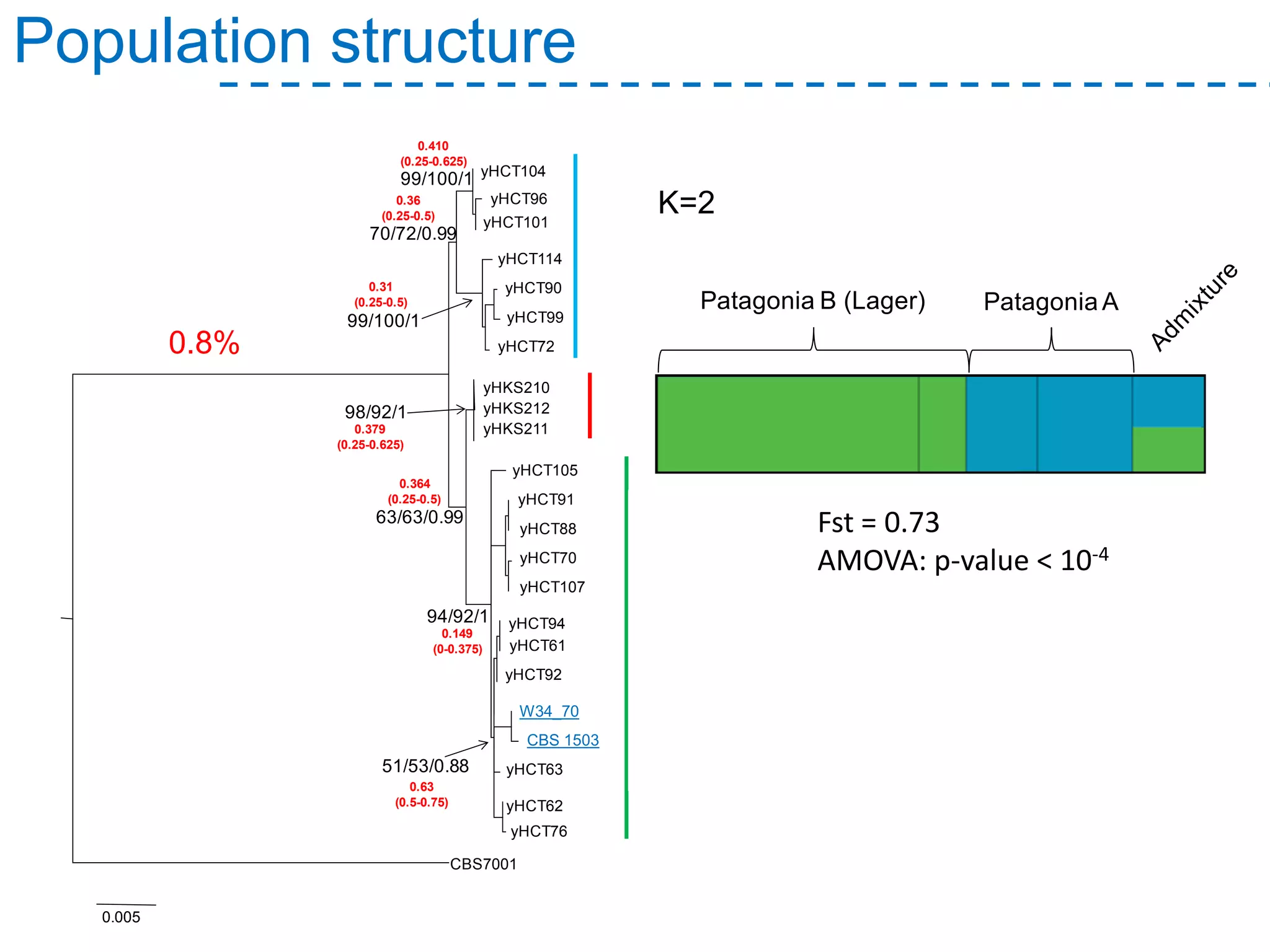
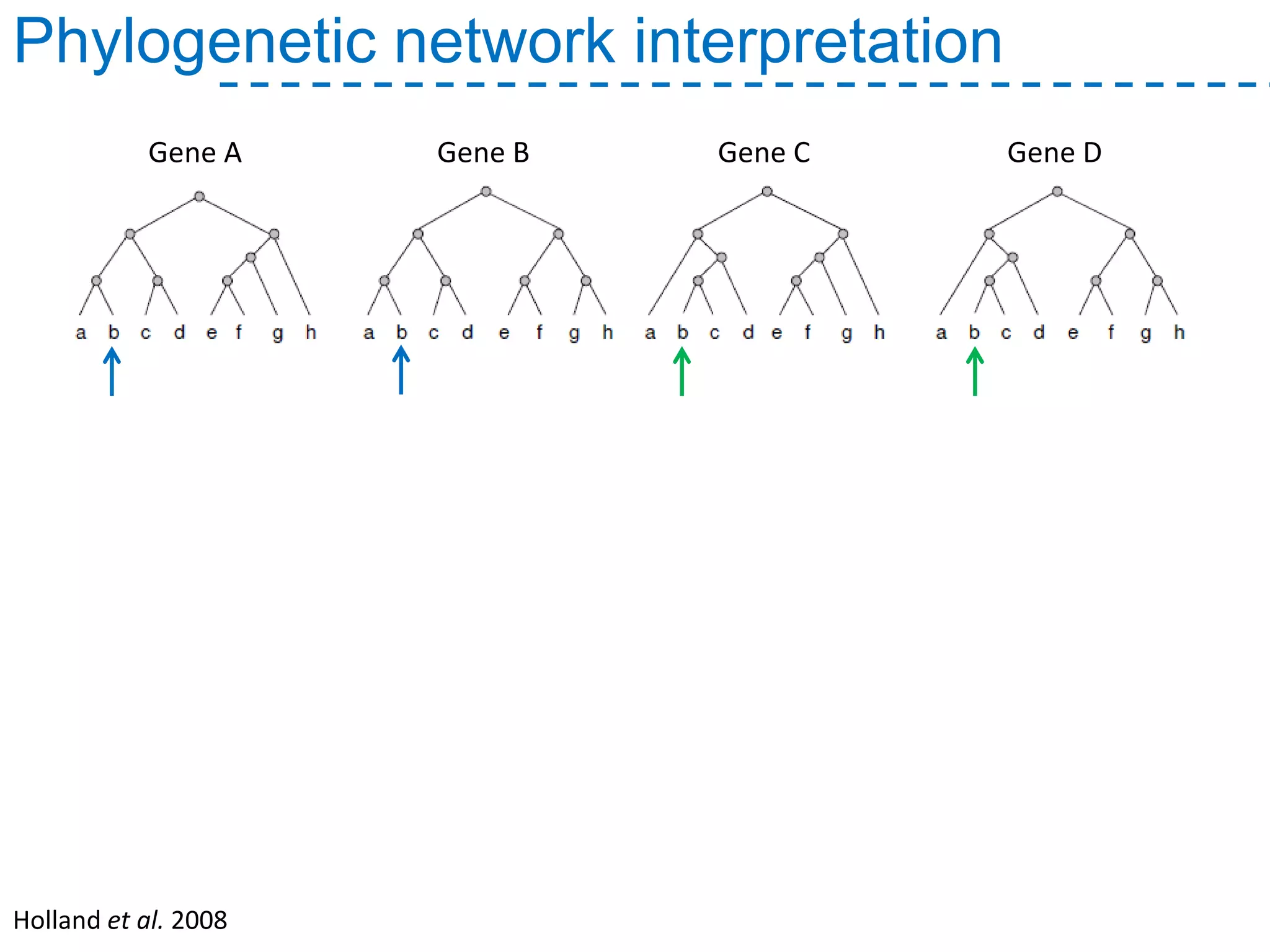
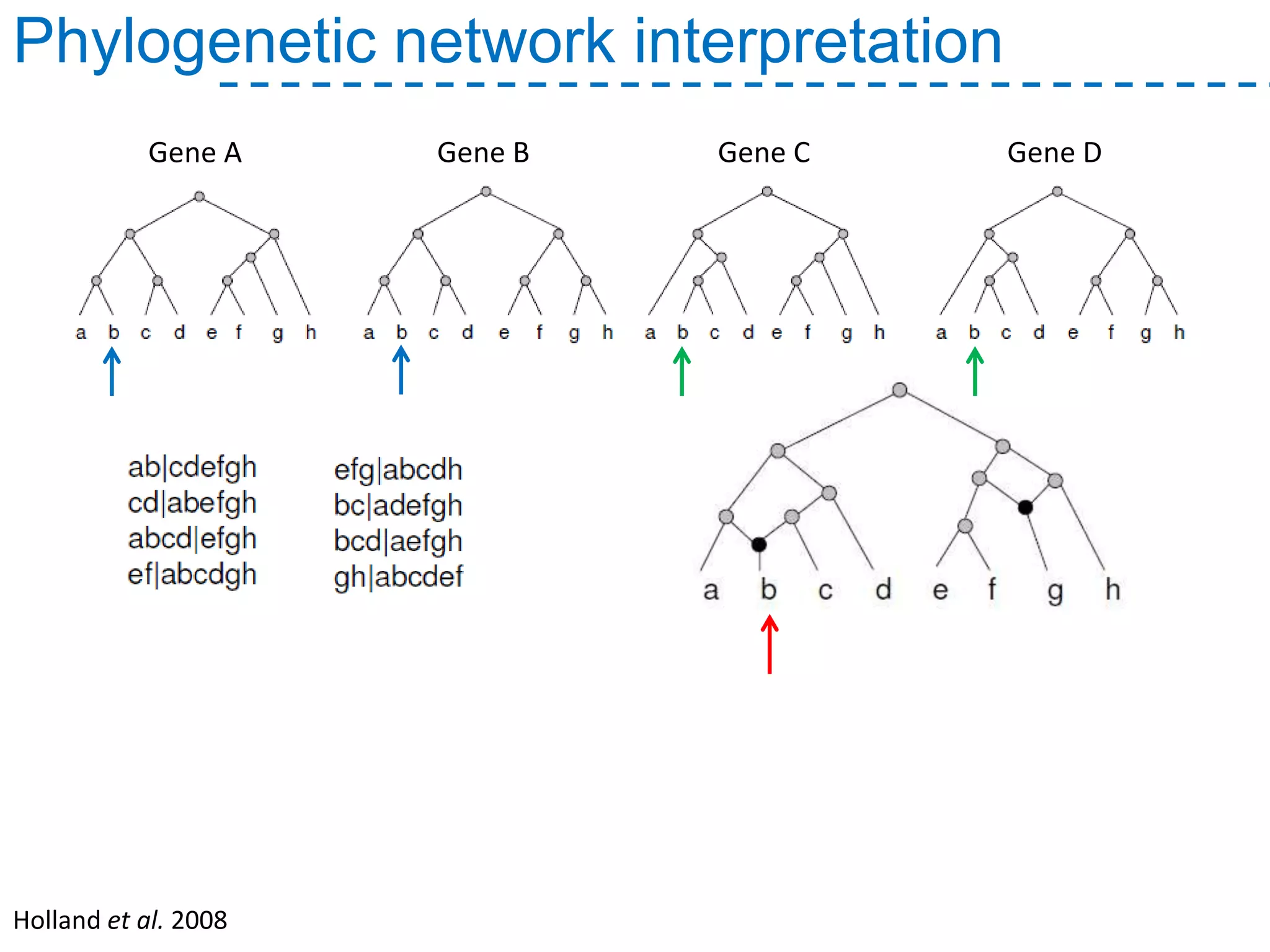
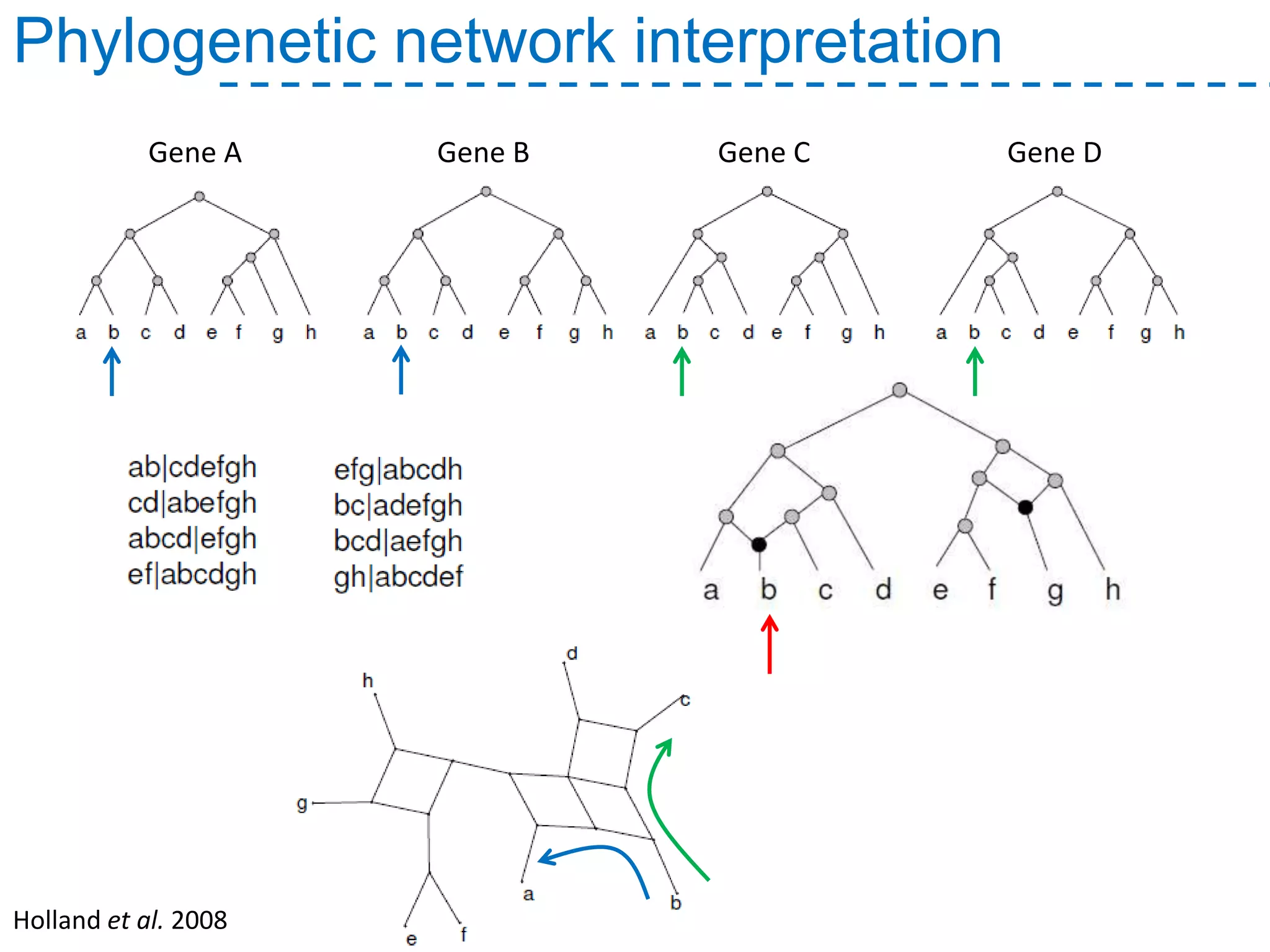
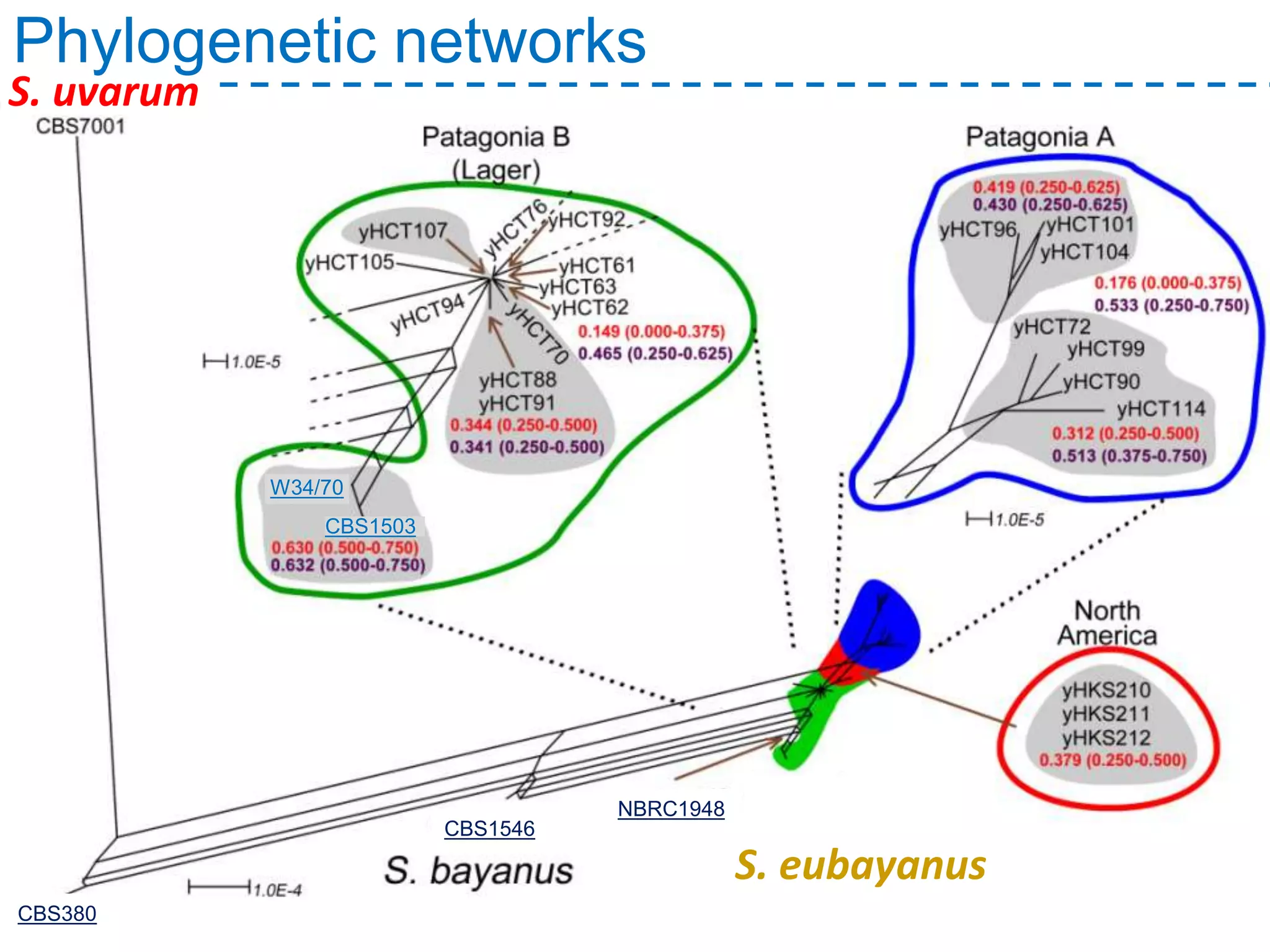
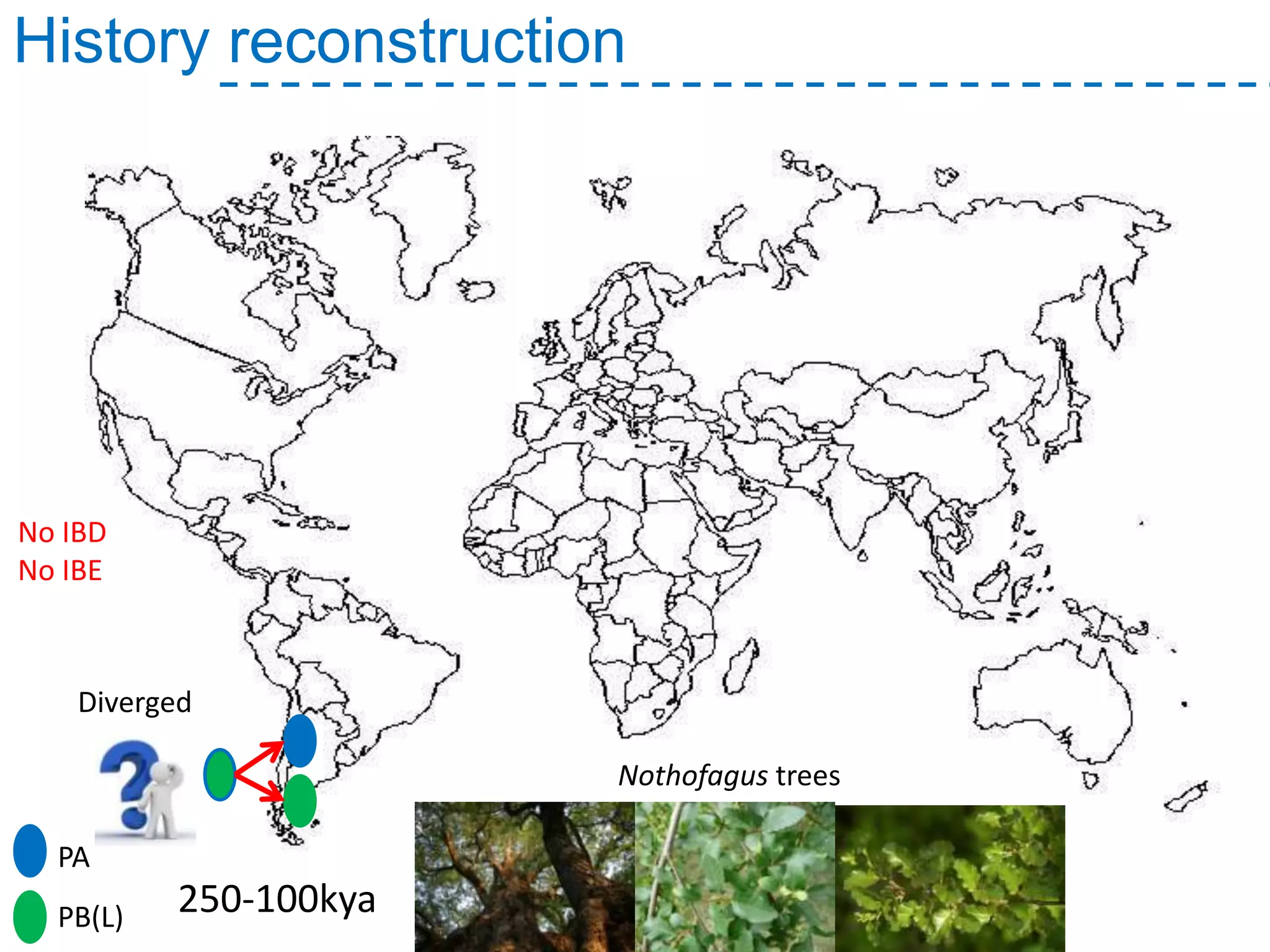
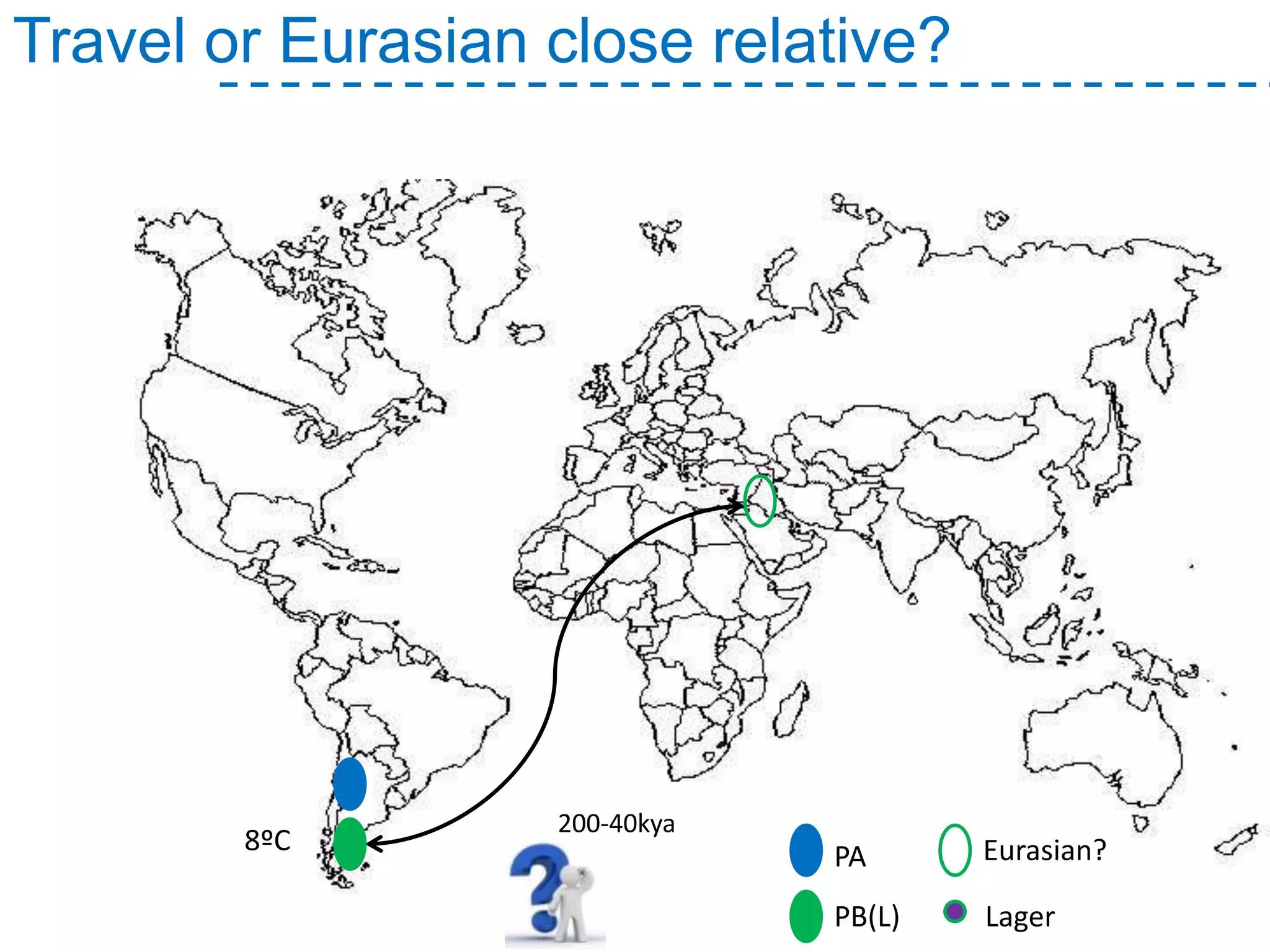
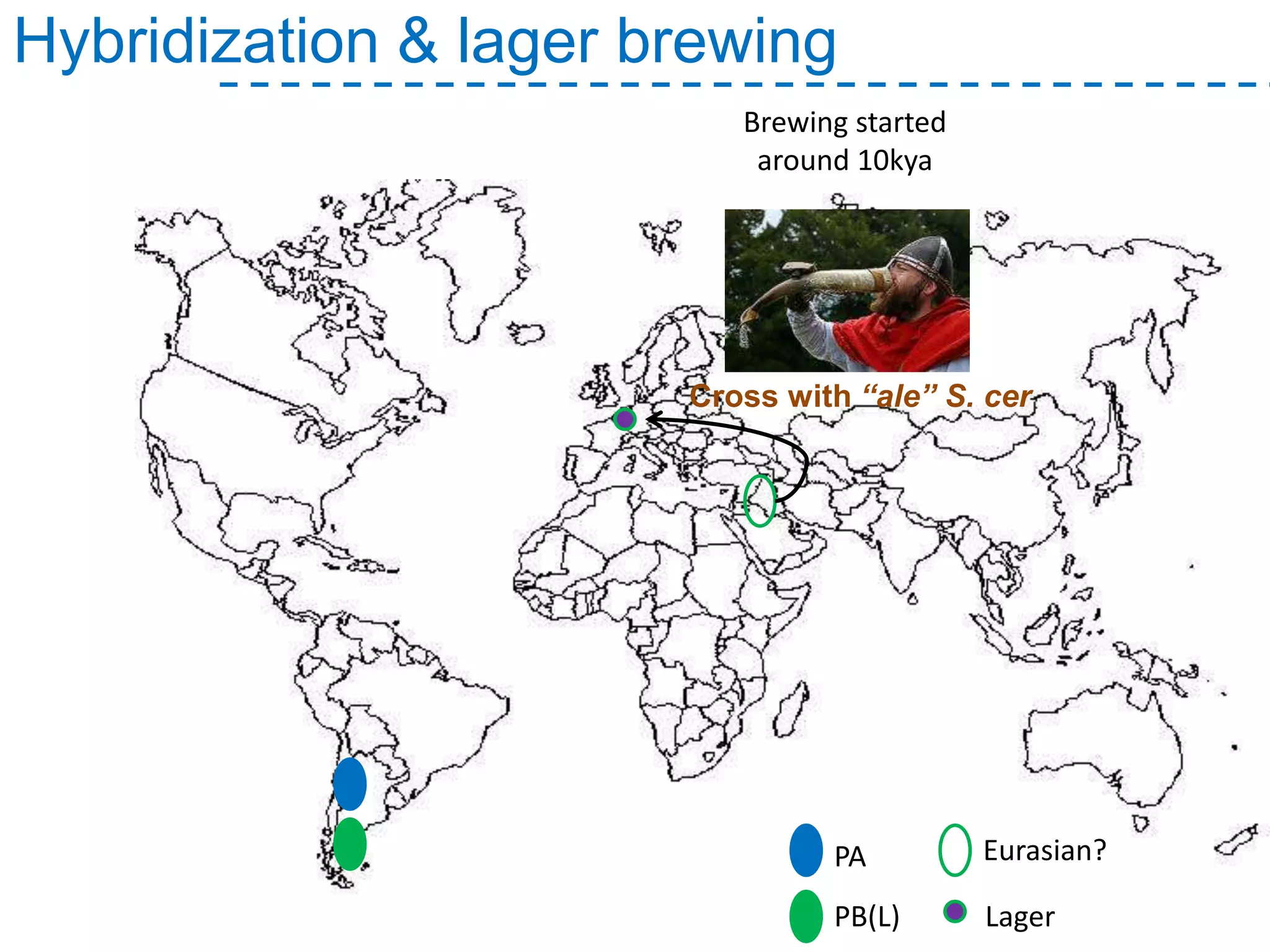
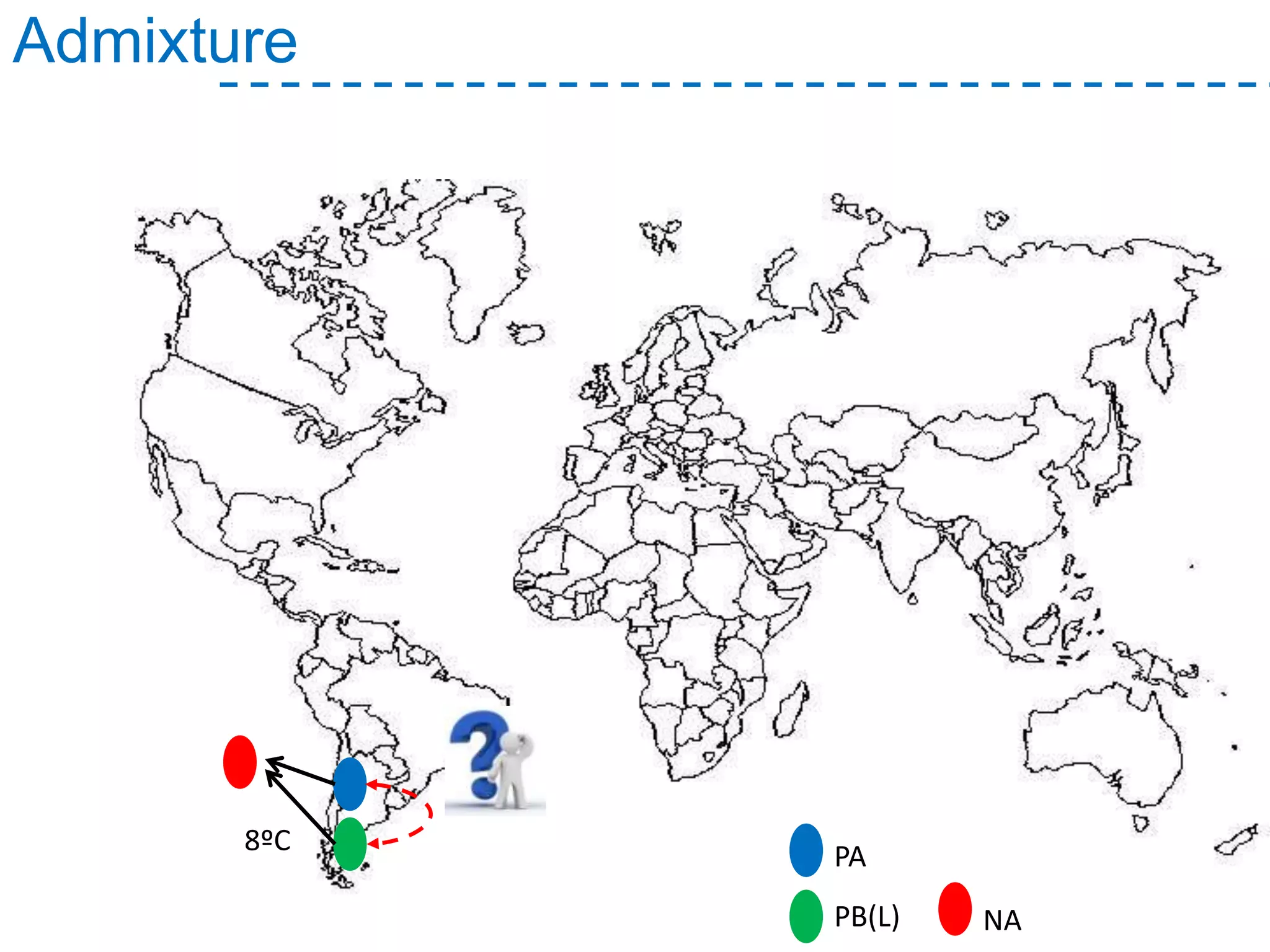
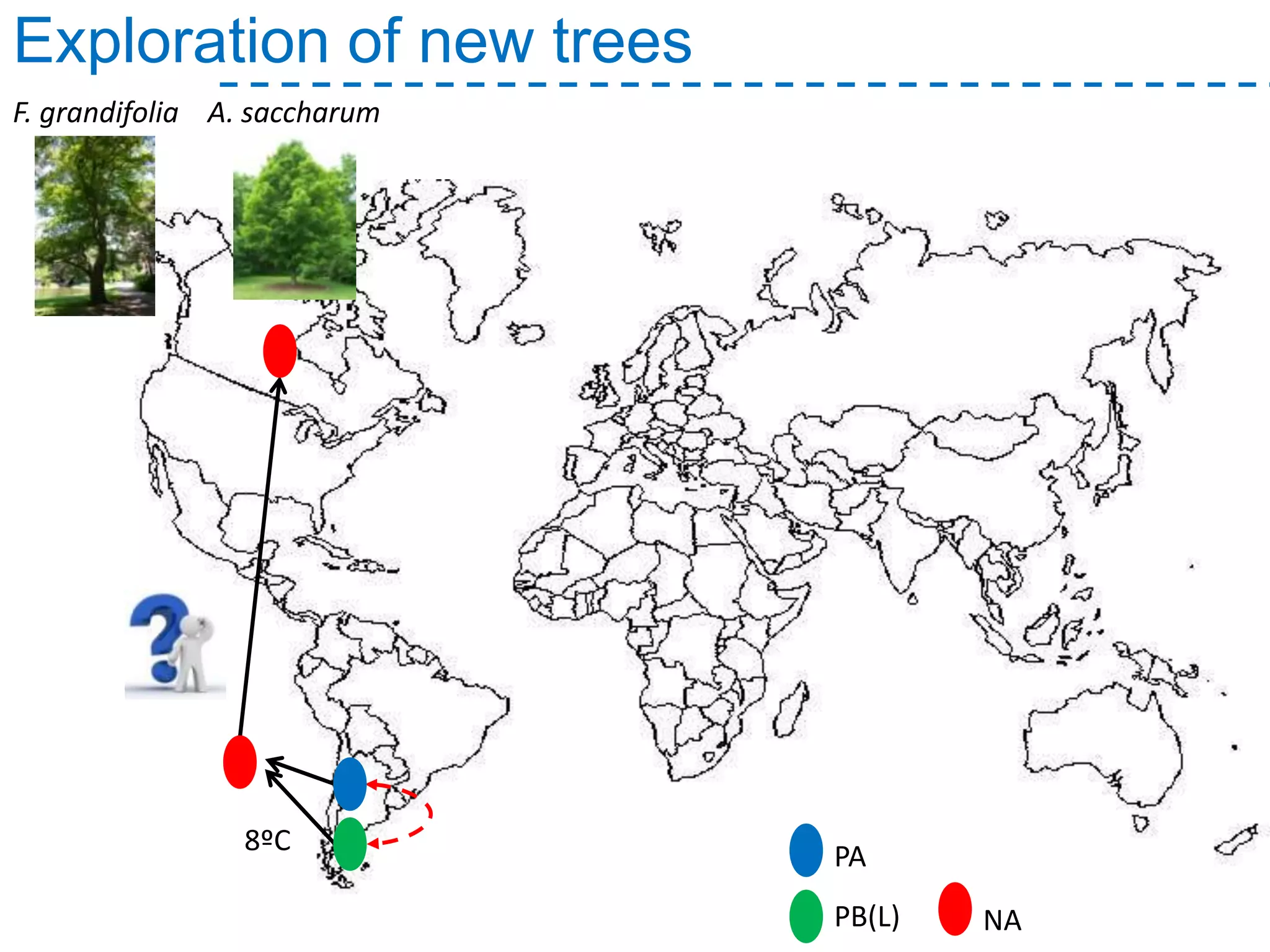
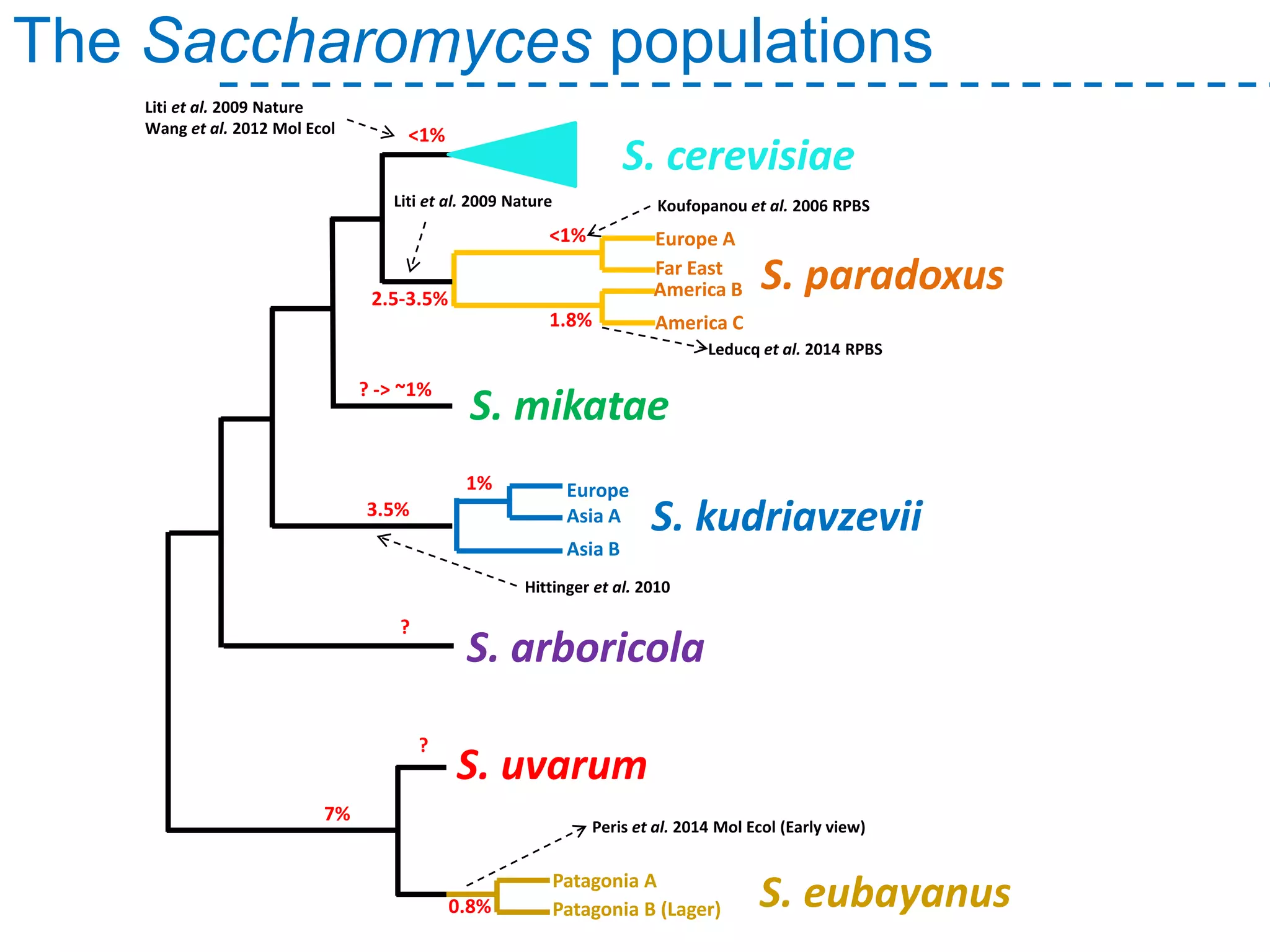
The document discusses the population structure and reticulate evolution of wild and brewing yeast, particularly focusing on the Saccharomyces genus and its diversity across different ecological environments. It raises key questions about hybridization, ecological factors driving differentiation, and the role of reticulate evolution in adaptation. The findings highlight the importance of reticulation events in yeast and the limited diversity of Saccharomyces eubayanus outside South America.