More Related Content
Similar to PosterPresentations.com-36x48-Template-V5 (2015_11_30 16_40_32 UTC) (20)
PosterPresentations.com-36x48-Template-V5 (2015_11_30 16_40_32 UTC)
- 1. RESEARCH POSTER PRESENTATION DESIGN © 2015
www.PosterPresentations.com
The regulation of arabinose operon in Escherichia Coli,
and the physical and regulatory properties of the operon’s
controlling gene, araC has been investigated for a while.
The ara operon is regulated by the AraC protein. Without
arabinose, the dimer AraC protein represses the structural
gene by binding and forms a loop, which prevents RNA
polymerase from binding to the promoter of the ara
operon, thereby blocking transcription. At a finer level,
however, despite the fact that it is one of the well-studied
regulatory proteins, much remains unknown about AraC.
Here we investigated the positive feedback of AraC
systems and found the bistable range through the
reflection of GFP expression, which is crucial for future
work. Combining synthetic biology and mathematical
modeling, this work sheds light on the bistable behaviors
emerging from sugar metabolism, which could be
exploited for biotechnology and therapeutics.
ABSTRACT
OBJECTIVES
Strains, Growth conditions, and media
All cloning experiments were performed in Escherichia coli
DH10B (Invitrogen), and measurements of positive feedback
response were conducted in DH10B and MG 1655. Cells were
grown at 37°C (unless specified) in liquid and solid Luria-
Bertani (LB) broth medium with 100 ug/ml ampicillin.
Plasmid construction
The receiver ParaC—AraC was constructed from six BioBrick
standard biological parts: Promoter activated by AraC in
concert with Arabinose, B ribosome binding site, RBS, AraC
gene, transcriptional terminator, and GFP reporter.
MATERIALS&METHODS
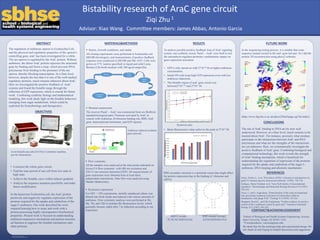
To analyze possible positive feedback loop of AraC signaling
system, one synthetic circuit, ParaC—AraC were built to test
the autoinducer-regulator-promoter combinations impact on
gene expression activation.
• GFP is only turned on with 2*10-10 M or higher arabinose
concentration.
• Initial ON cells keep high GFP expression even with low
arabinose inductions.
• The bistable region of araC gene circuit was
between2*10^-10 and 2*10-7 M.
RESULTS FUTURE WORK
In the sequencing testing process, it is notable that some
sequence mutant existed in the araC gene tail part. So I did the
protein 3D reconstruction using phyre2 software.
(http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index)
The site of AraC binding to DNA are by now well
understood. However, at a finer level, much remains to be
learned about AraC. For instance, precisely what residues
participate in the interactions between AraC and RNA
polymerase and what are the strengths of the interactions
are yet unknown. Here, we systematically investigate the
positive feedback of AraC gene. Combining biological and
computational technology, this work reveals the strength
of AraC binding mechanism, which is beneficial for
understanding the regulation of expression of the proteins
required for the uptake and catabolism of the sugar L-
arabinose, DNA looping and allosteric mechanism.
REFERENCES
Emini, Emilio A., et al. "Prevention of HIV-1 infection in chimpanzees by
gpl20 V3 domain-specific monoclonal antibody." (1992): 728-730.
Gallegos, Maria-Trinidad, et al. "Arac/XylS family of transcriptional
regulators." Microbiology and Molecular Biology Reviews 61.4 (1997):
393-410.
Gross, J., and E. Englesberg. "Determination of the order of mutational
sites governing l-arabinose utilization in Escherichia coliBr by
transduction with phage P1bt." Virology 9.3 (1959): 314-331.
Sheppard, David E., and Ellis Englesberg. "Further evidence for positive
control of the L-arabinose system by gene araC." Journal of molecular
biology 25.3 (1967): 443-454
. CONTANCT&ACKNOWLEDGEMENT
1School of Biological and Health Systems Engineering, Arizona
State University, Tempe, AZ 85287, USA
*Correspondence: xiaowang@asu.edu
We thank Hao for the technique help and experimental design. We
also thank Qi and Fuqing for helpful discussions and suggestions
• Construct the whole gene circuit.
• Find the time period of one cell from low state to
high state.
• Achieve the bistable curve within inducer gradient.
• Analyze the sequence mutation possibility and make
future modification.
In the bacterium Escherichia coli, the AraC protein
positively and negatively regulates expression of the
proteins required for the uptake and catabolism of the
sugar L-arabinose. Our work described the work
required learning how to assay and work with a
protein possessing highly uncooperative biochemical
properties. Present work is focused on understanding
arabinose-responsive mechanism and protein structure
ad function to engineer the bistable mechanism onto
other proteins.
Advisor: Xiao Wang. Committee members: James Abbas, Antonio Garcia
Ziqi Zhu1
Bistability research of AraC gene circuit
Arabinose induced synthetic
gene network
Flow cytometry
All the samples were analyzed at the time points indicated on
Accuri C6 flow cytometer with 488 nm excitation and
530+15 nm emission detection (GFP). All measurements of
gene expression were obtained from at least three
independent experiments. Date files were analyzed using
Matlab (Mathworks).
Hysteresis experiment
For OFF ON experiments, initially uninduced culture was
diluted into fresh media and induced with various amounts of
arabinose. Flow cytometry analyses were performed at 2hr,
4hr, 7hr, and 12hr to monitor the fluorescence levels, which
generally became stable after 7 hr induction according to our
experience.
Hysteresis plot
• Mean fluorescence value achieves the peak at 2*10-7 M.
RBS secondary structure is a potential reason that might affect
the protein expression due to the binding of ribosome and
mRNA.
B0032 (weak) RBS mutant (strong)
TCACACAGGAAAG AAAGAGGAGAAA
CONCLUSIONS
www.biotech.uiuc.edu (C6 Flow Cytometry machine,
just for illustration)