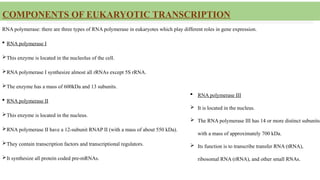
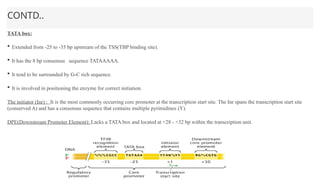
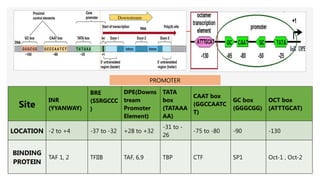
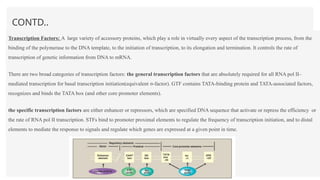
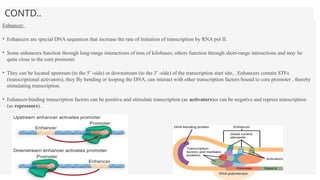
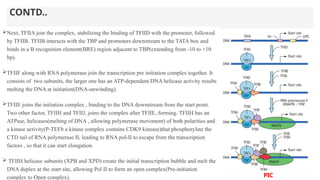
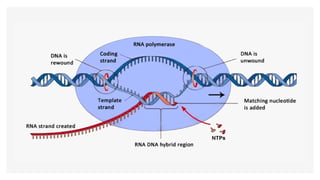
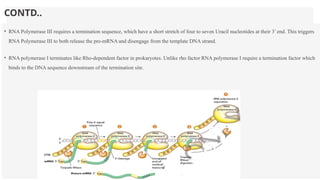
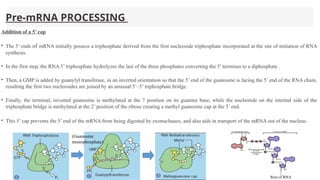
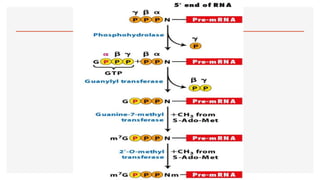
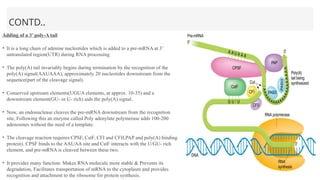
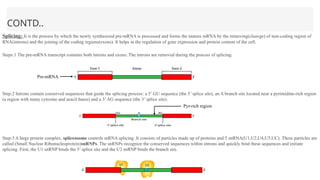
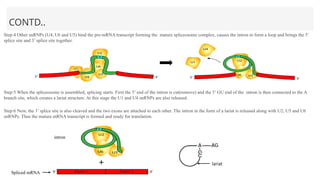
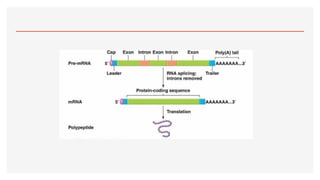
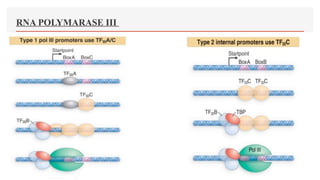
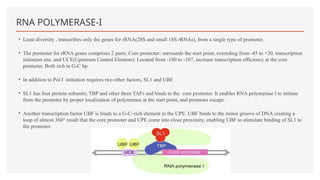
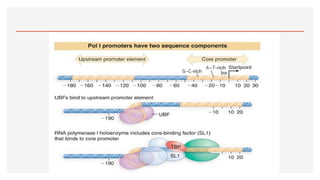
This presentation outlines the complex process of eukaryotic transcription, which involves separate RNA polymerases for synthesizing various RNA types and a variety of transcription factors that aid in initiating and regulating transcription. Key components discussed include the role of core and proximal promoters, enhancer sequences, and the mechanisms of initiation, elongation, and termination of transcription. Additionally, it details the processing of pre-mRNA through capping, polyadenylation, and splicing to produce mature mRNA ready for translation.