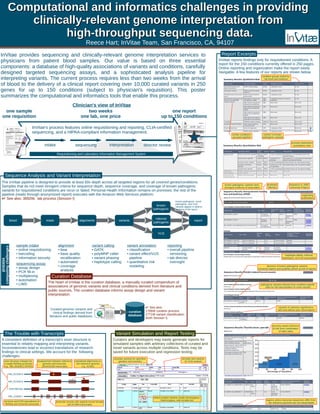
Invitae provides clinical genome interpretation services derived from high-throughput sequencing, producing detailed reports for up to 150 conditions within two weeks. Their system combines a curated database of pathogenic variants, rigorous sequencing protocols, and secure data management. The process emphasizes accuracy in variant interpretation while maintaining patient privacy through compliance with HIPAA standards.