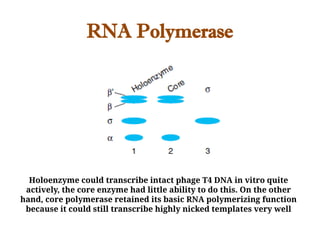
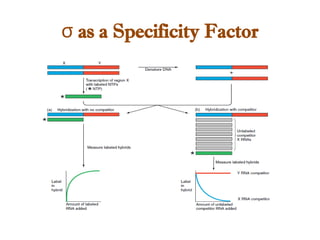
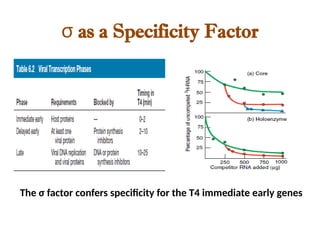
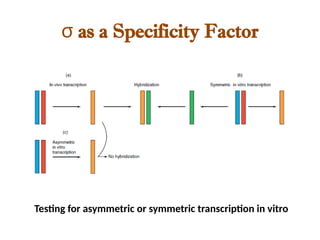
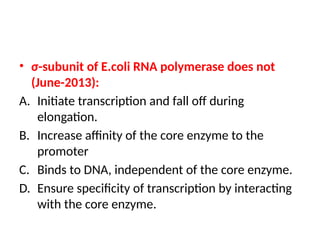
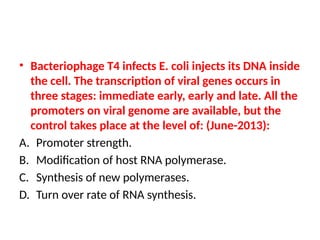
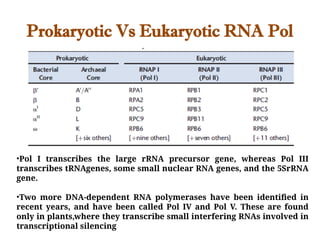
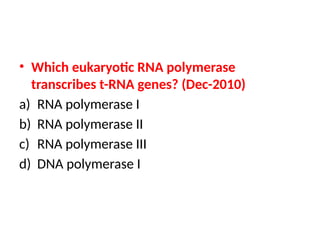
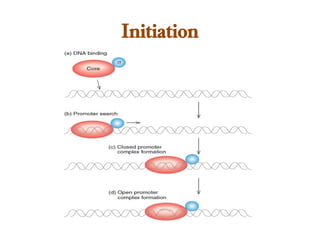
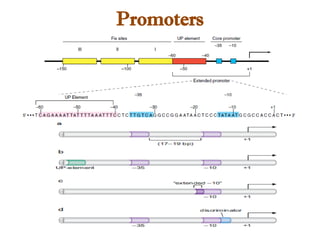
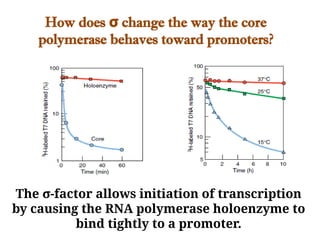
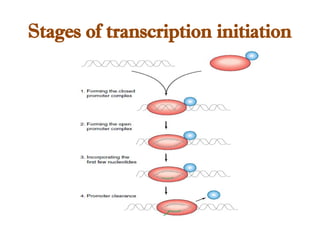
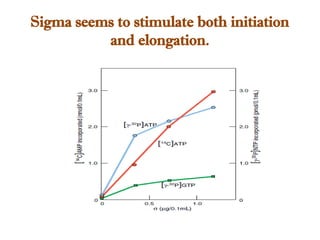
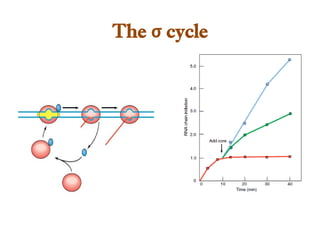
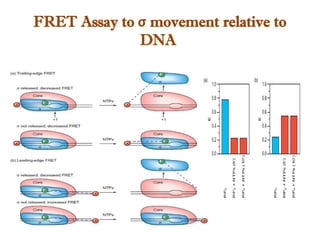
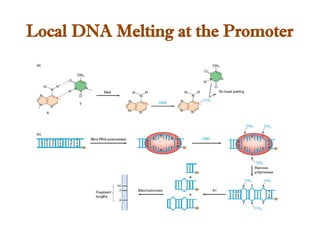
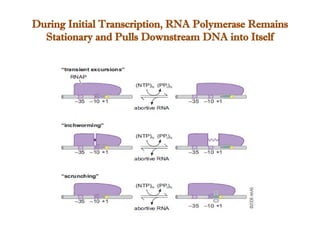
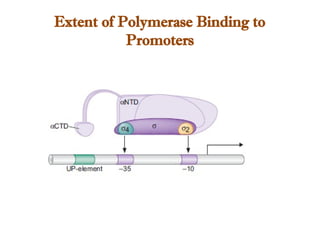
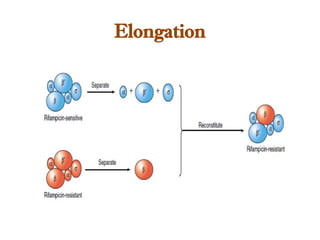
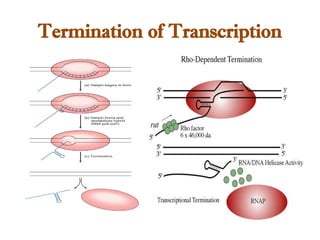
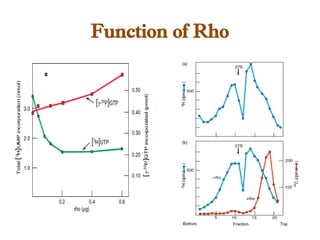
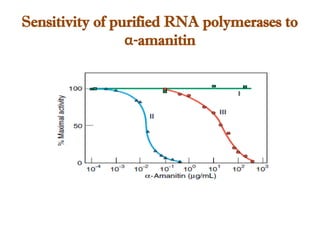
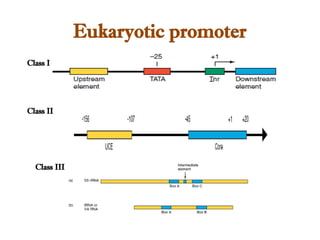
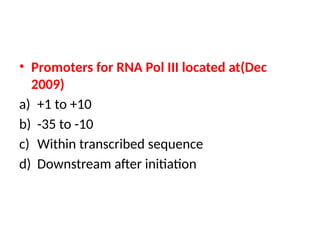
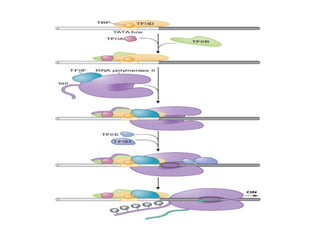
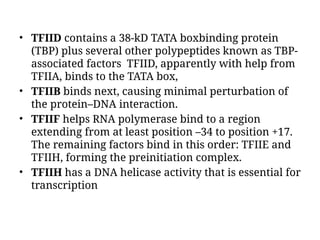
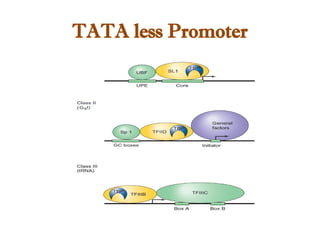
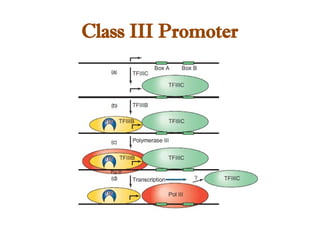
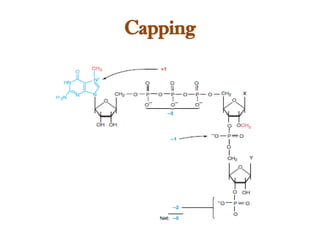
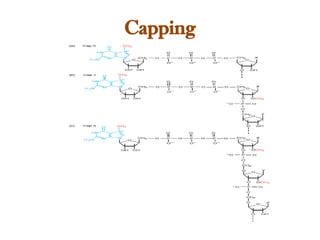
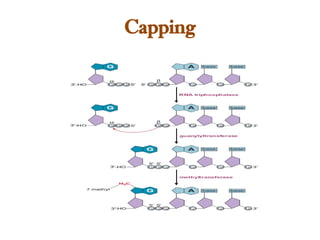
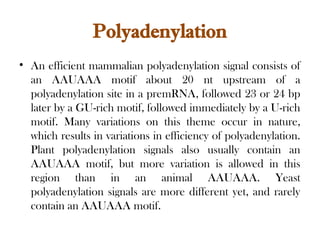
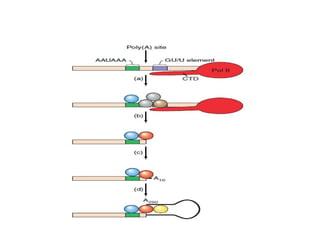
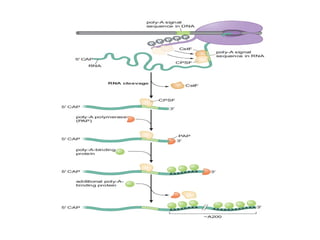
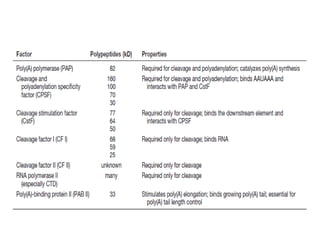
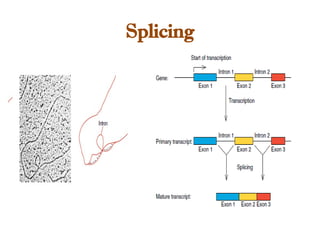
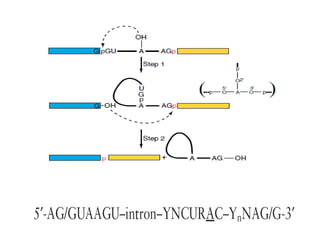
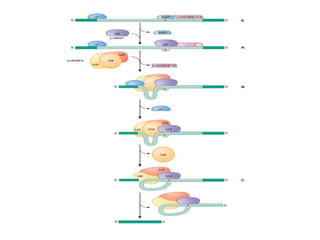
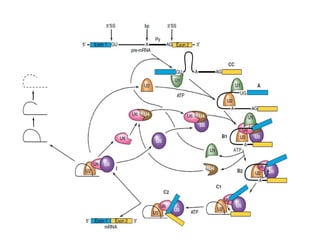
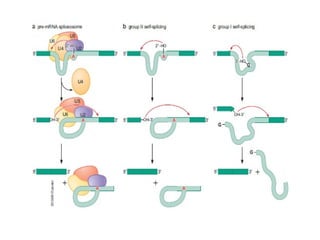
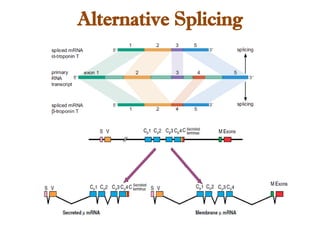
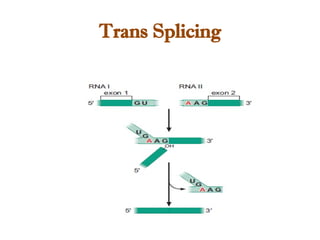
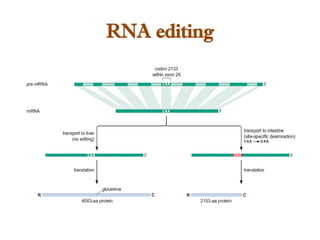
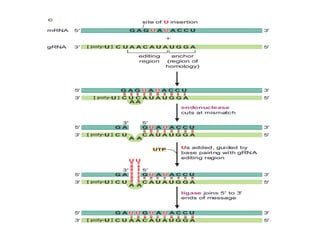
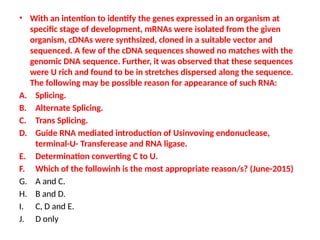
The document discusses RNA synthesis and processing, covering topics such as RNA polymerases, transcription factors, and various stages of transcription initiation and termination. It highlights the differences between prokaryotic and eukaryotic RNA polymerases, details processes like capping, polyadenylation, and splicing, and touches on DNA damage related to transcription. Additionally, it mentions specific RNA polymerase functions and the genetic implications of transcription factors.