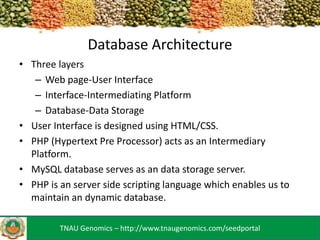
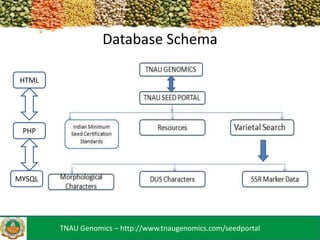
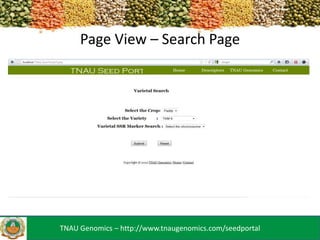
The TNAU Seed Portal is developed to provide a comprehensive and user-friendly database on seed standards, varietal characteristics, and quality certification, utilizing SSR marker fingerprinting to ascertain seed purity and classification. It includes a three-layer architecture with an HTML/CSS user interface, PHP as an intermediary, and MySQL for data storage, facilitating dynamic updates. Future developments aim to enhance accessibility through mobile applications and expand the SSR marker data available.