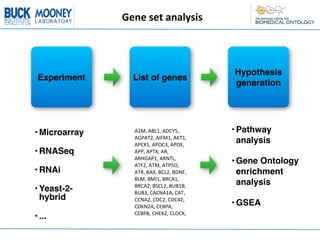
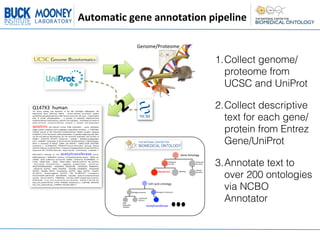
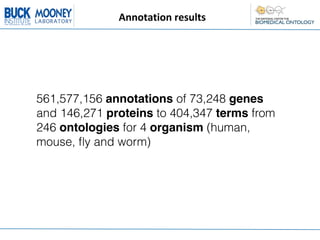
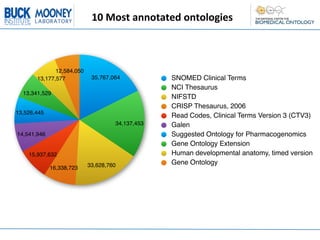
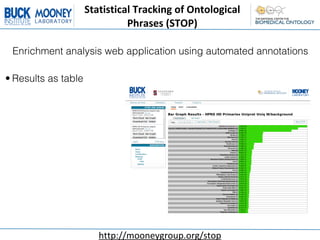
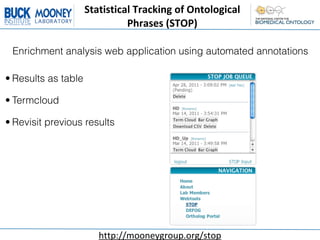
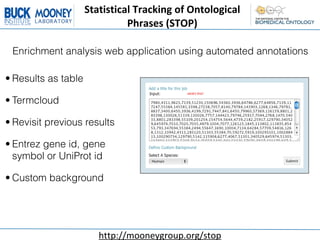
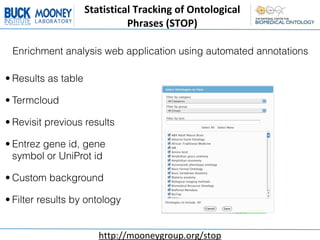
This document describes an automatic gene annotation pipeline that uses descriptive text about genes and proteins to annotate them to terms in over 200 ontologies via the NCBO annotator service. The pipeline involves:
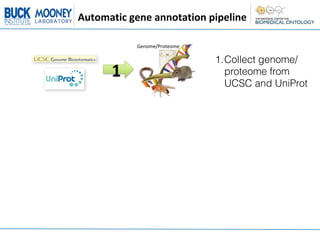
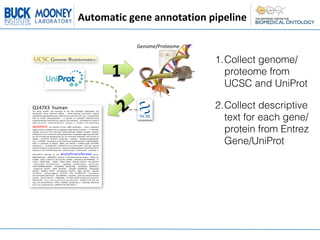
1. Collecting the genome/proteome from databases and extracting descriptive text for each gene/protein from Entrez Gene and UniProt.
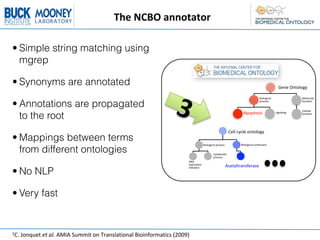
2. Annotating the extracted text to terms in over 200 ontologies using the NCBO annotator service.
3. This allows genes/proteins to be annotated to a wide range of ontologies, not just Gene Ontology, leveraging up-to-date information in existing gene/protein databases.