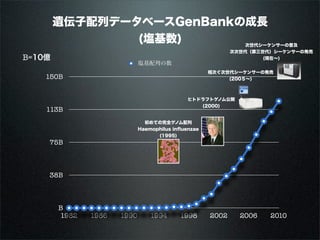
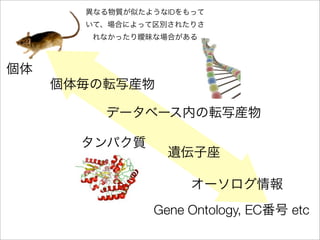
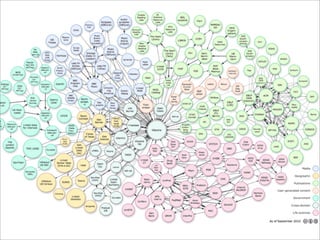
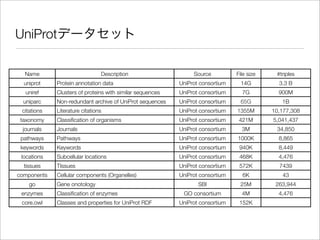
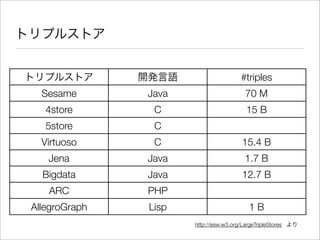
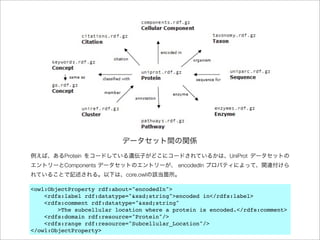
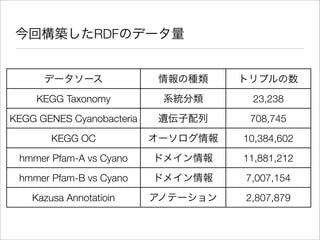
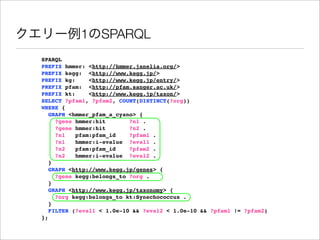
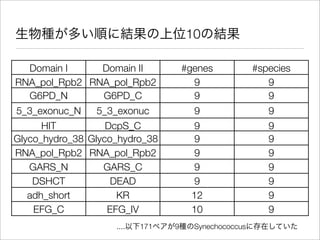
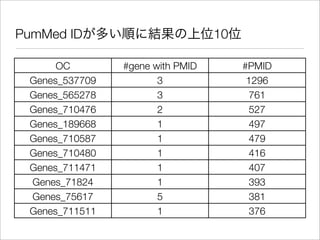
The document discusses using Resource Description Framework (RDF) to represent biological data from sources like UniProt, PDBJ, and KEGG. It provides examples of SPARQL queries that can be used to query and analyze relationships within and across RDF datasets, such as finding gene families present in multiple cyanobacterial species or linking genes to biological pathways and literature citations. Large public RDF datasets are described along with Semantic Web technologies for publishing and querying linked open data.