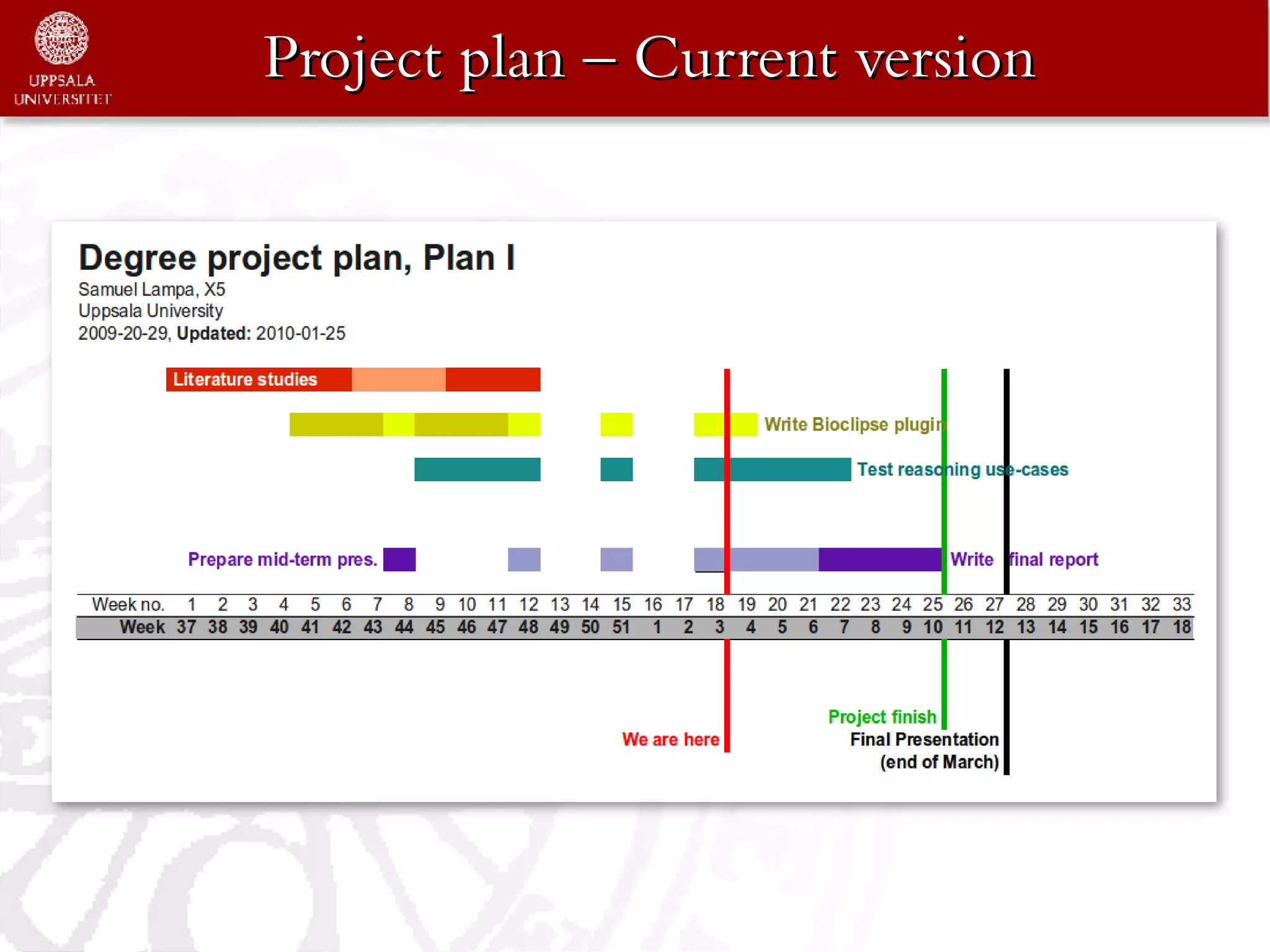
The document is a status report on a degree project that integrates blipkit/bioprolog for semantic reasoning in Bioclipse. It compares two reasoners, Pellet and Blipkit, focusing on biochemical questions formulated as Prolog queries in terms of speed and expressiveness. The project has successfully integrated Blipkit, conducted initial performance testing, and outlines further tasks including NMR spectrum similarity search and further comparisons between Prolog and Pellet.










![Prolog code example
% === SOME FACTS ===
hasHBondDonors( substanceX, 3 ). % “substance X has 3 H-bond donors”
% etc …
% === A RULE ("RULE OF FIVE" ÀLA PROLOG) ===
Head Implication (“If [body] then [head]”)
isDrugLike( Substance ) :-
hasHBondDonorsCount( Substance, HBDonors ),
HBDonors <= 5,
hasHBondAcceptorsCount( Substance, HBAcceptors ),
HBAcceptors <= 10,
hasMolecularWeight( Substance, MW ),
MW < 500.
Body
% === QUERYING THE RULE ===
?- isDrugLike(substanceX) Comma means conjunction (“and”)
true.
?- isDrugLike(X)
X = substanceX ;
X = substanceY.
Capitalized terms are always variables](https://image.slidesharecdn.com/presentationfinal-12700663977349-phpapp01/75/2nd-Proj-Update-Integrating-SWI-Prolog-for-Semantic-Reasoning-in-Bioclipse-13-2048.jpg)







![Example Bioclipse/Prolog script
blipkit.init();
blipkit.loadRDFToProlog("nmrshiftdata.100.rdf.xml");
// Define a “convenience prolog method”
blipkit.loadPrologCode("
hasPeak( Subject, Predicate ) :-
rdf_db:rdf( Subject,
'http://www.nmrshiftdb.org/onto#hasPeak',
Predicate ).
");
// Call the convenience method (which in turn executes it's
// “body”), and returns all mathing results as an array
var resultList =
blipkit.queryProlog(["hasPeak","10","Subject","Predicate"]);](https://image.slidesharecdn.com/presentationfinal-12700663977349-phpapp01/75/2nd-Proj-Update-Integrating-SWI-Prolog-for-Semantic-Reasoning-in-Bioclipse-22-2048.jpg)
![Example Bioclipse/Prolog script
blipkit.init();
blipkit.loadRDFToProlog("nmrshiftdata.100.rdf.xml");
// Define a “convenience prolog method”
blipkit.loadPrologCode("
hasPeak( Subject, Predicate ) :-
rdf_db:rdf( Subject,
'http://www.nmrshiftdb.org/onto#hasPeak',
Predicate ).
"); Prolog rule to load into prolog engine
// Call the convenience method (which in turn executes it's
// “body”), and returns all mathing results as an array
var resultList =
blipkit.queryProlog(["hasPeak","10","Subject","Predicate"]);
Prolog method to call
Limit the number of results Prolog variables](https://image.slidesharecdn.com/presentationfinal-12700663977349-phpapp01/75/2nd-Proj-Update-Integrating-SWI-Prolog-for-Semantic-Reasoning-in-Bioclipse-23-2048.jpg)