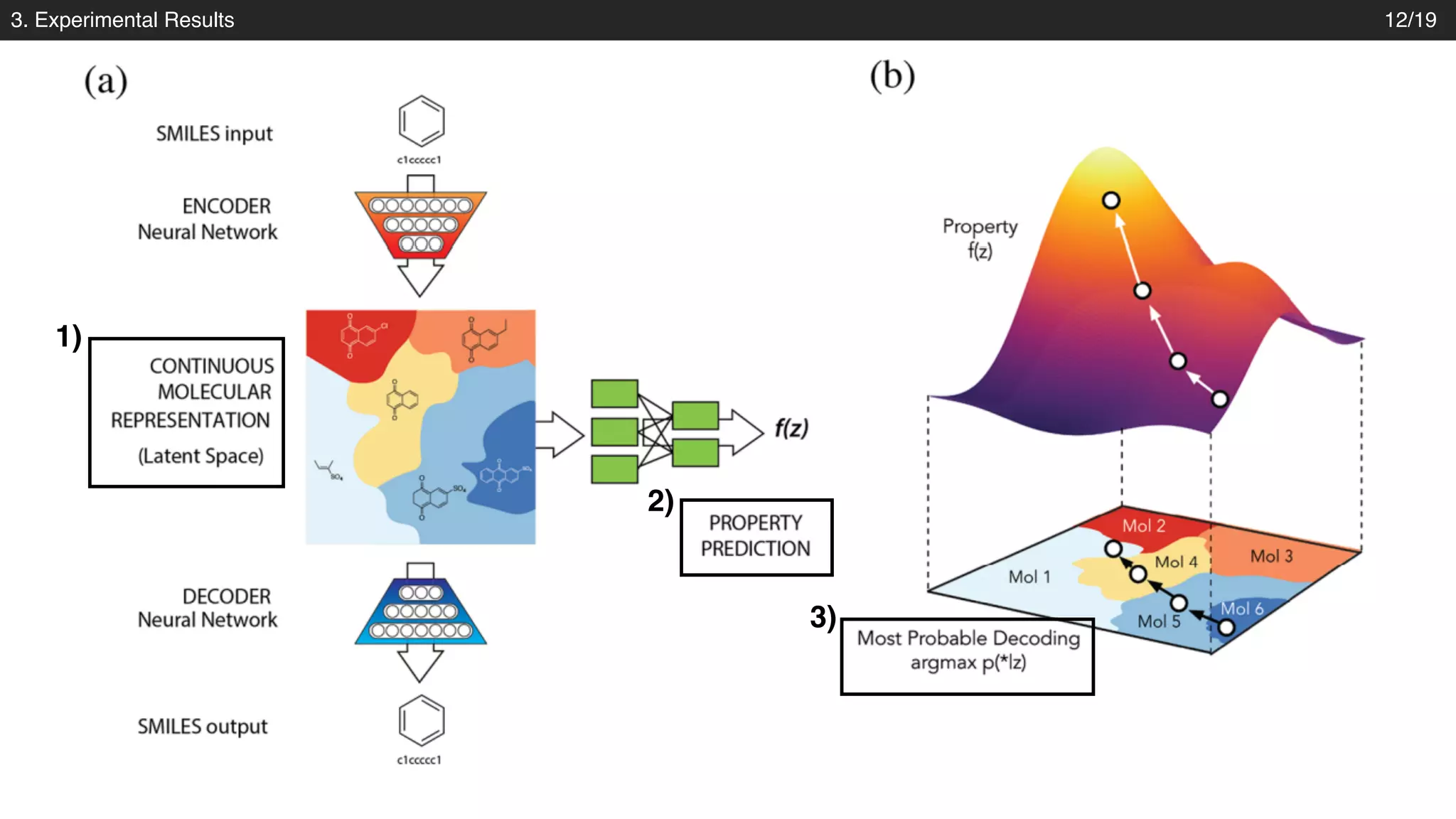
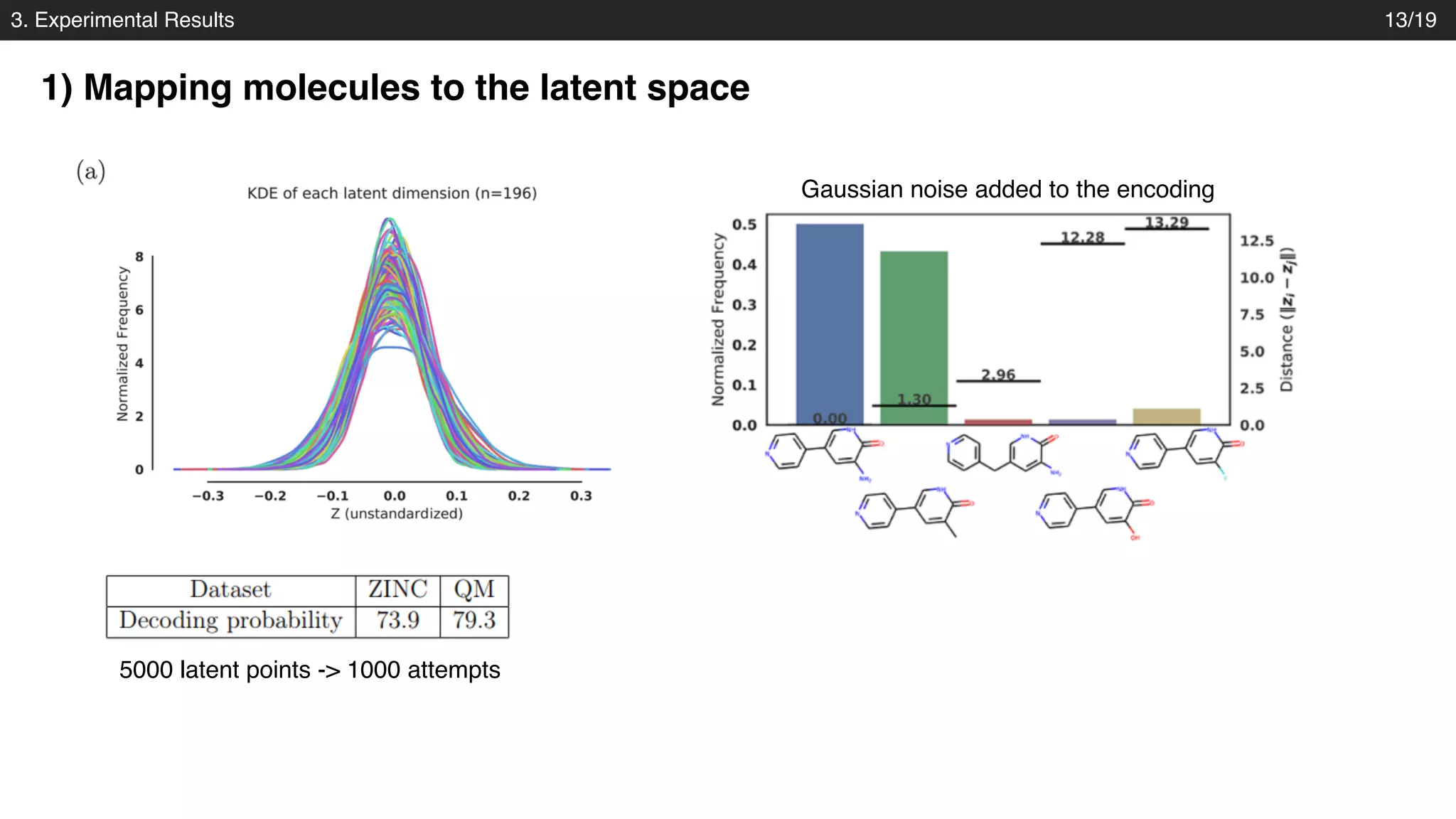
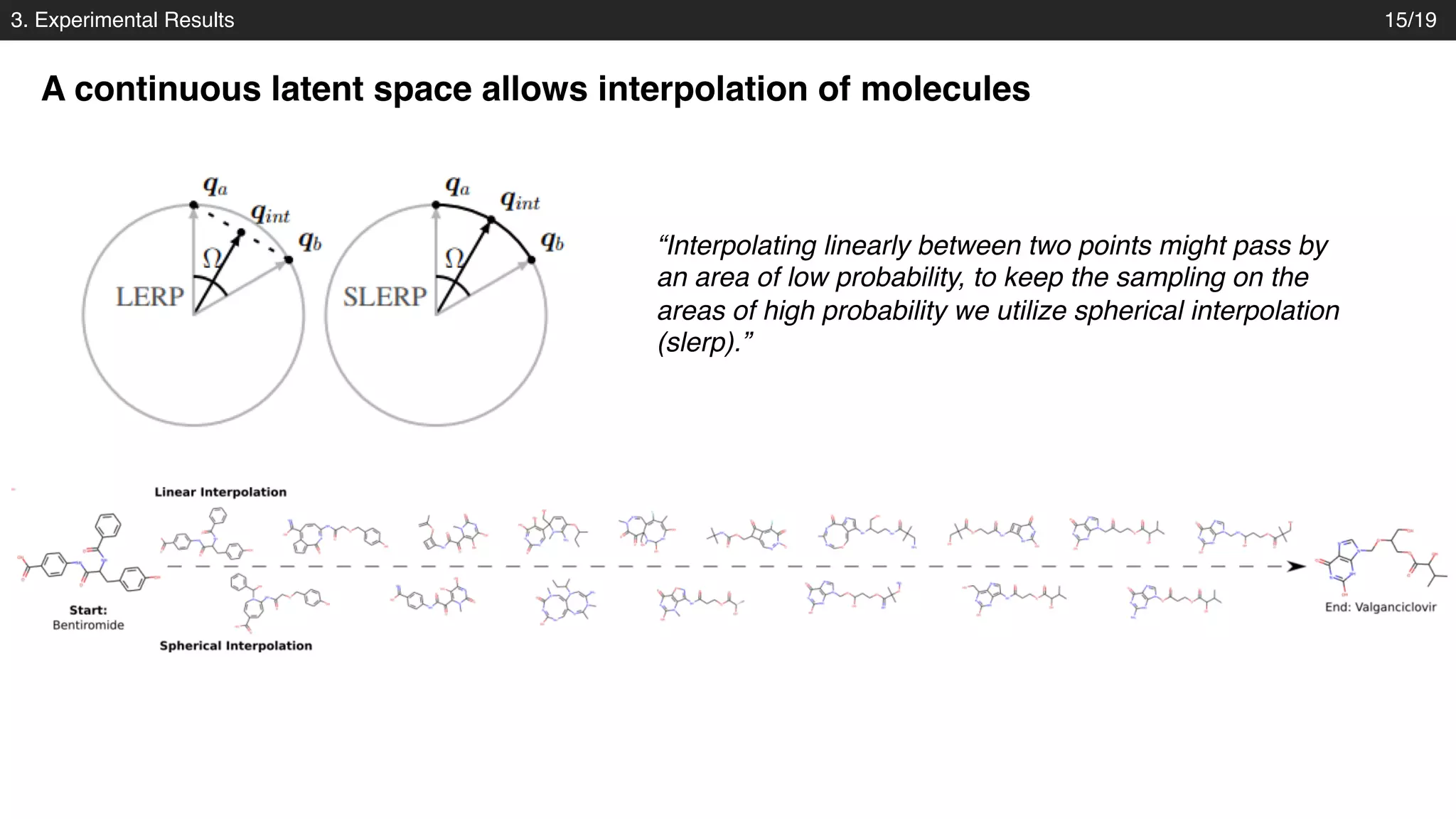
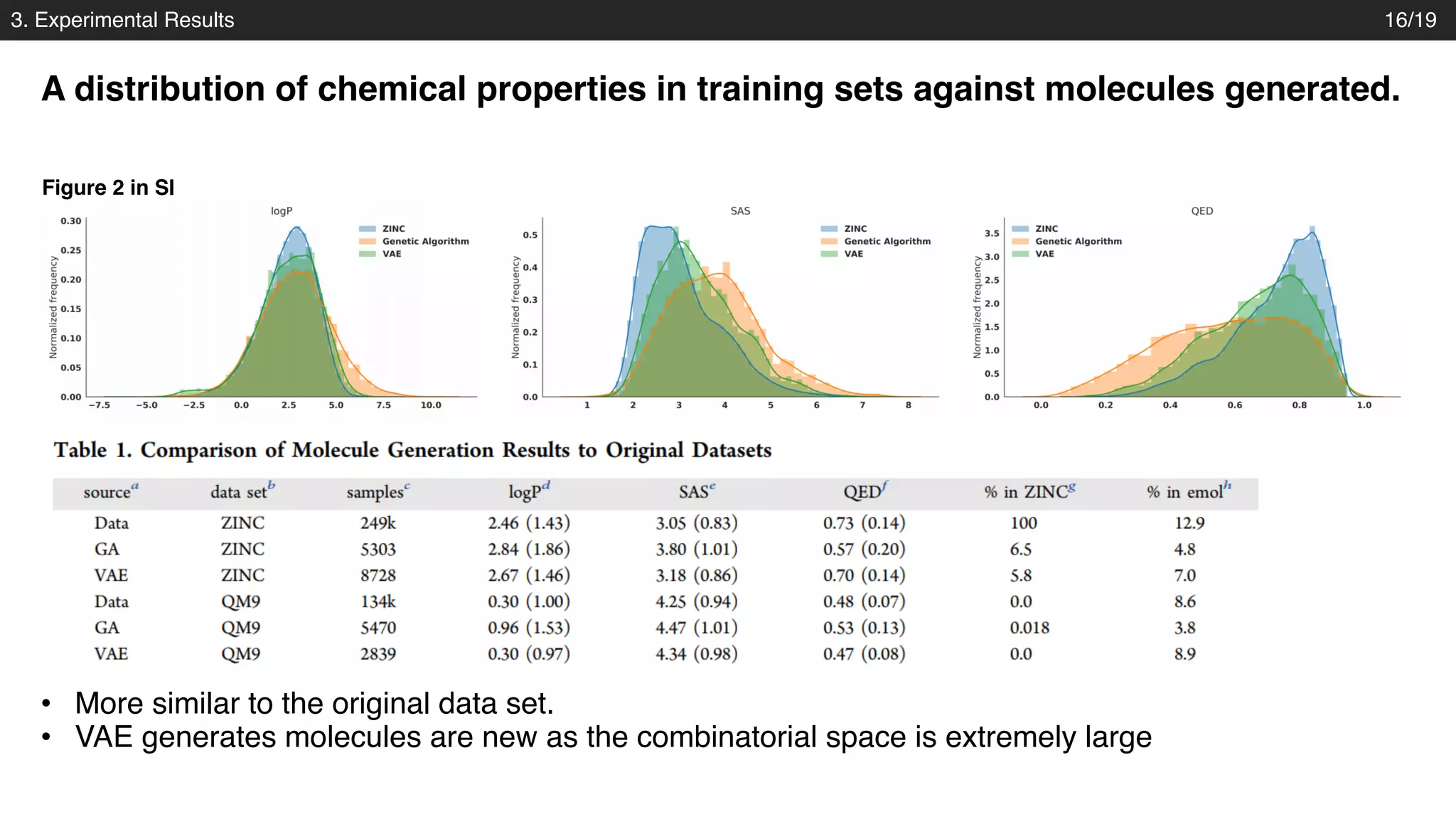
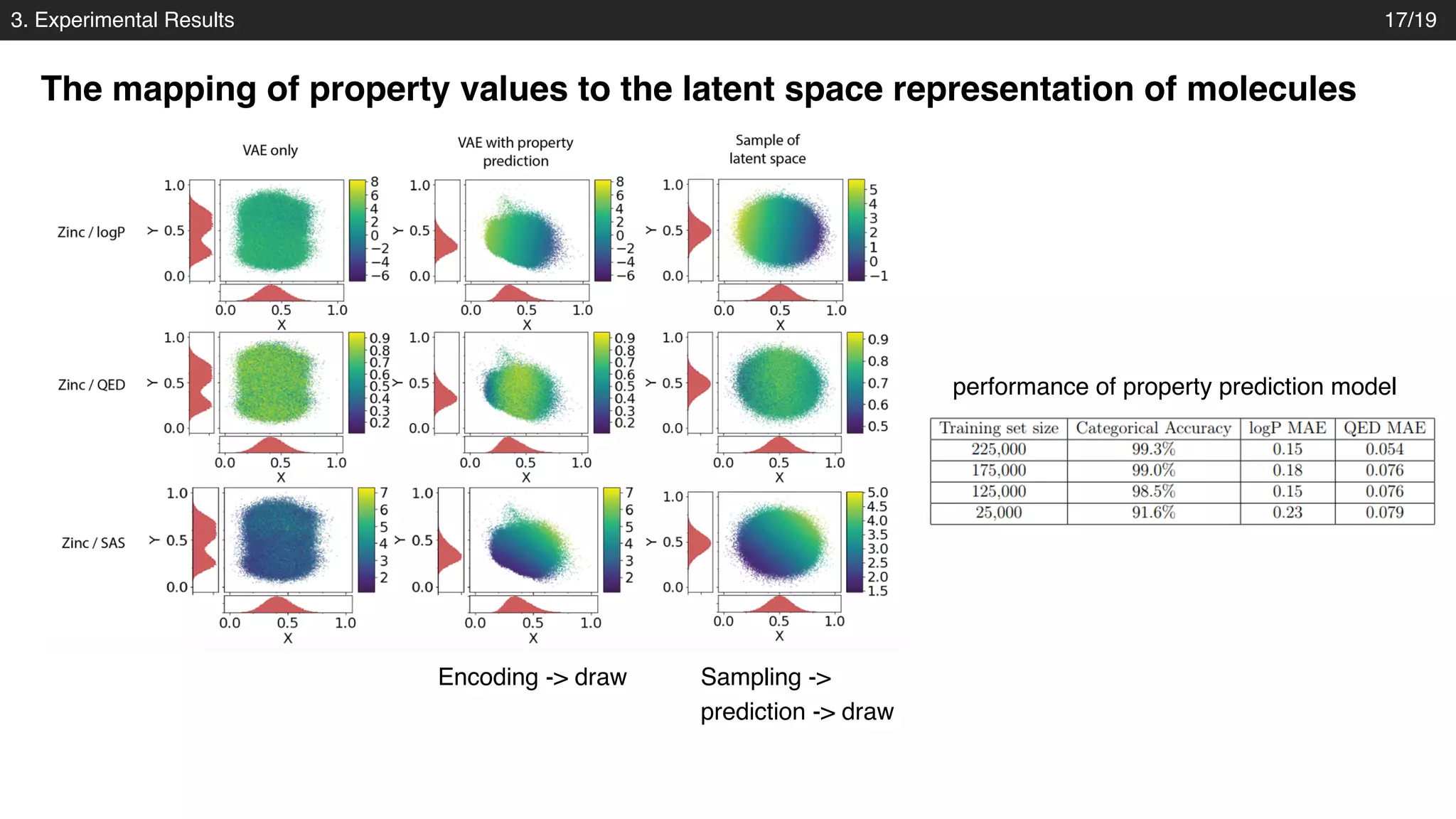
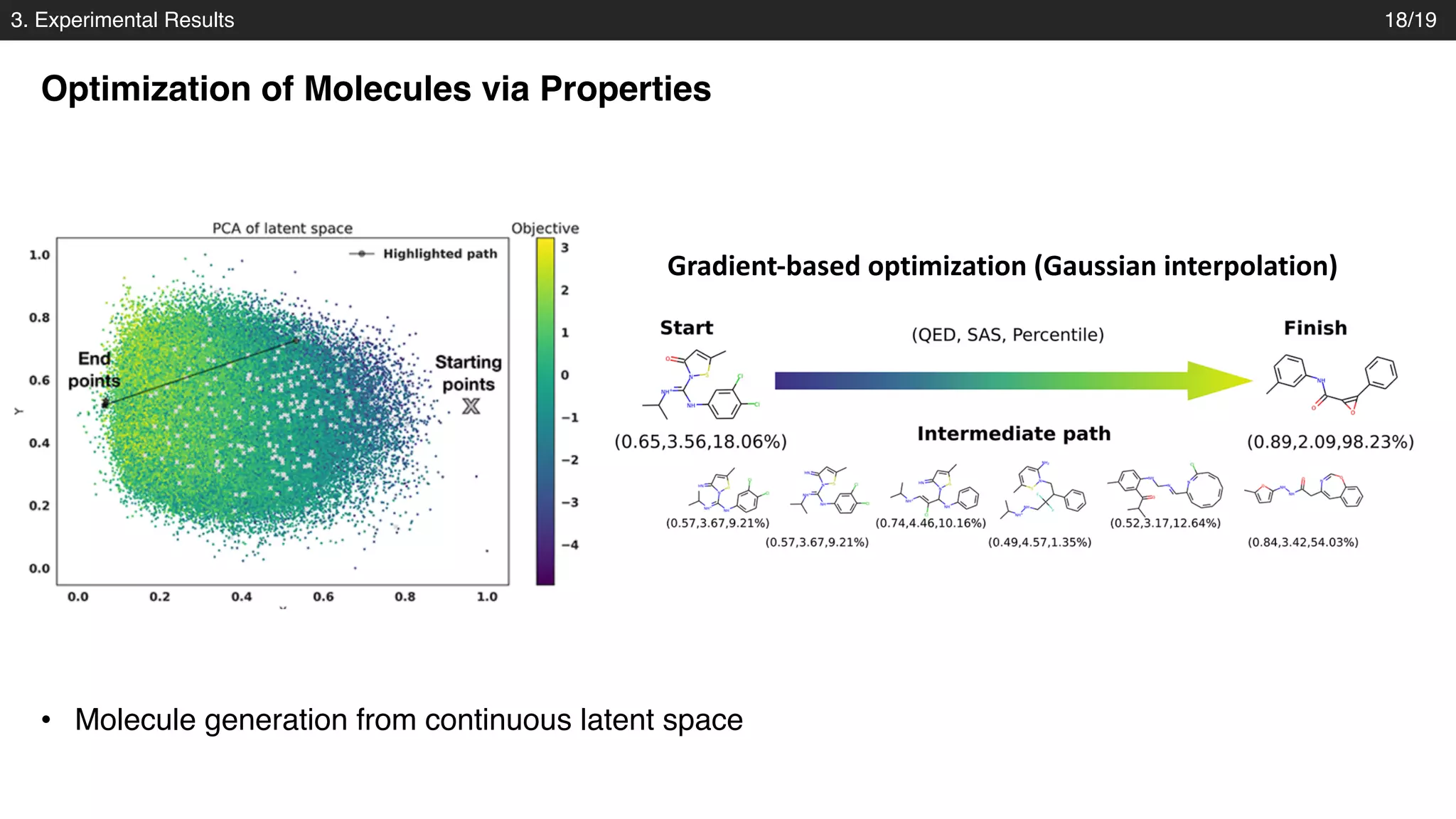
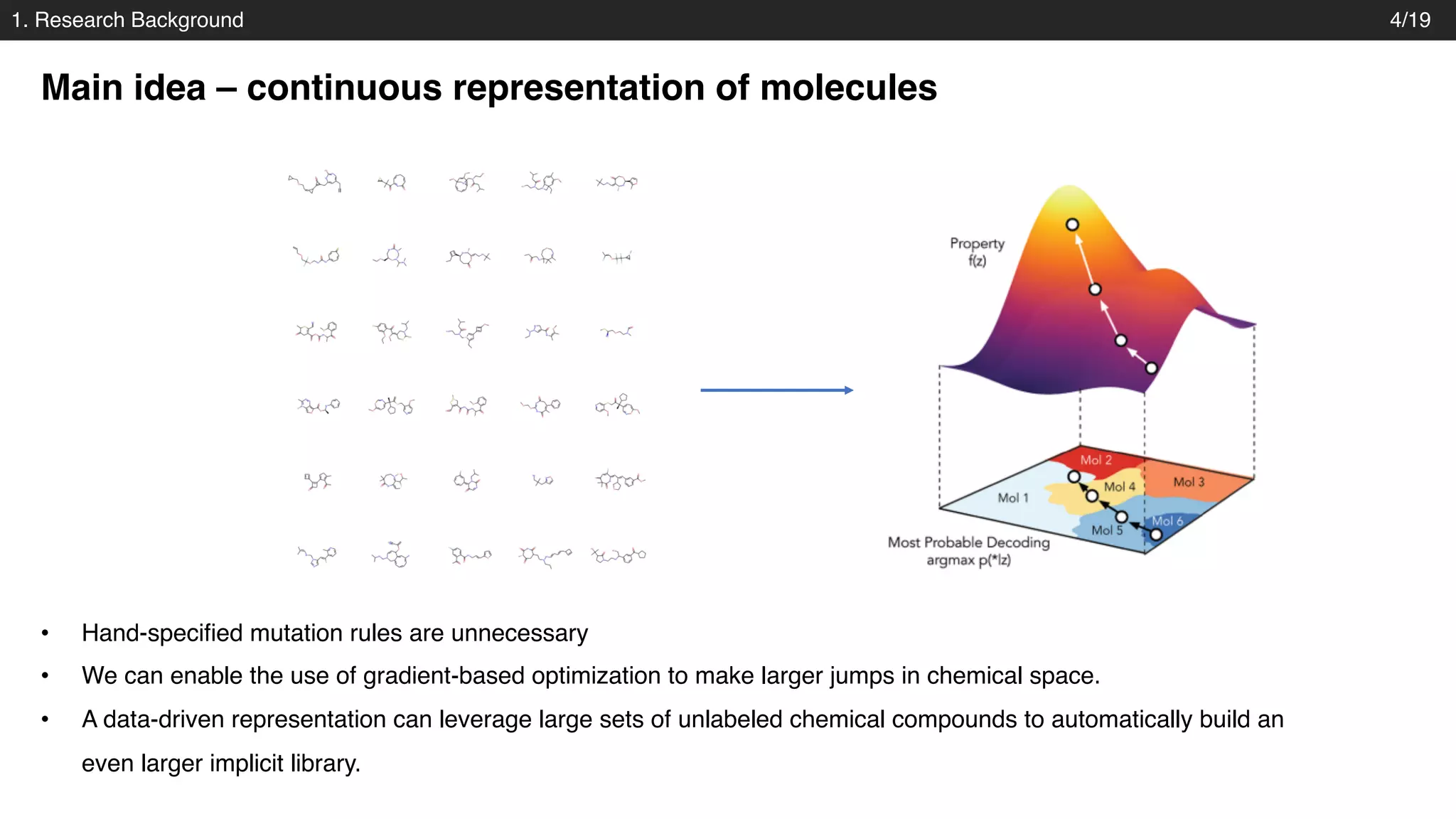
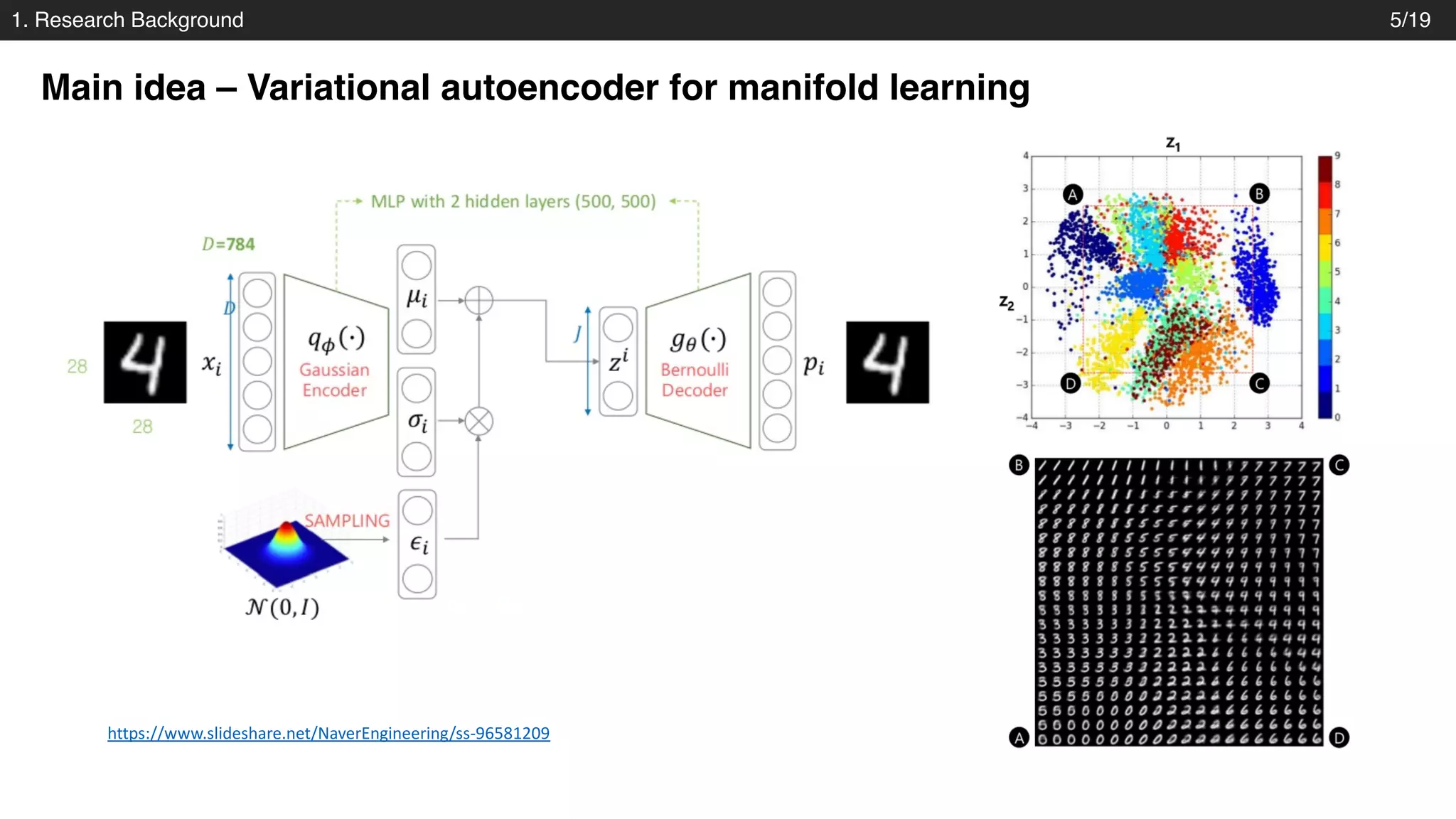
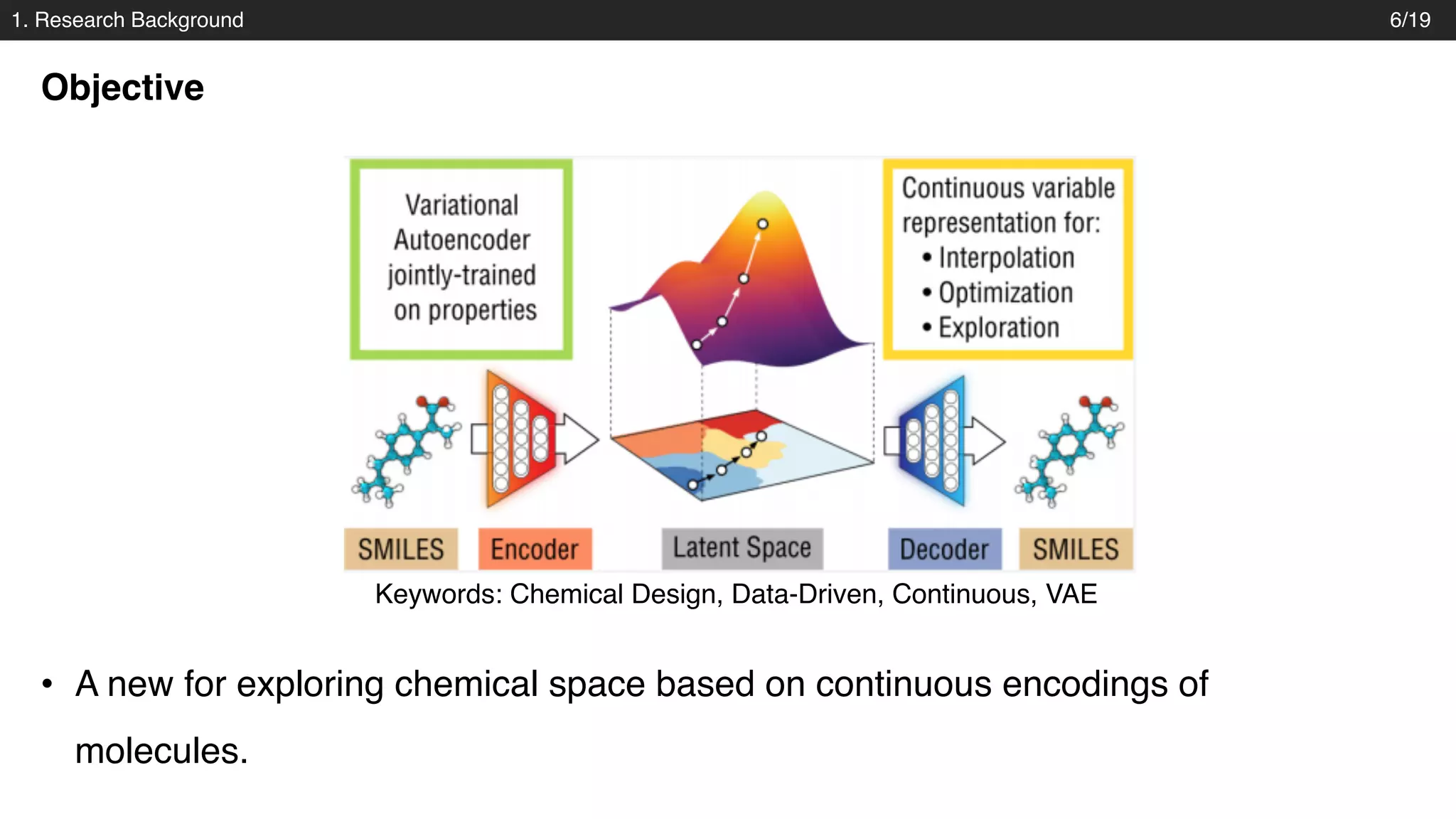
The document discusses a novel approach to exploring chemical space using continuous encodings of molecules through variational autoencoders (VAEs). It highlights the integration of data-driven techniques and gradient-based optimization methods for drug and material design, leveraging large databases of drug-like molecules. The approach shows promise in generating new molecules with desirable properties while optimizing various objective functions in molecular design.



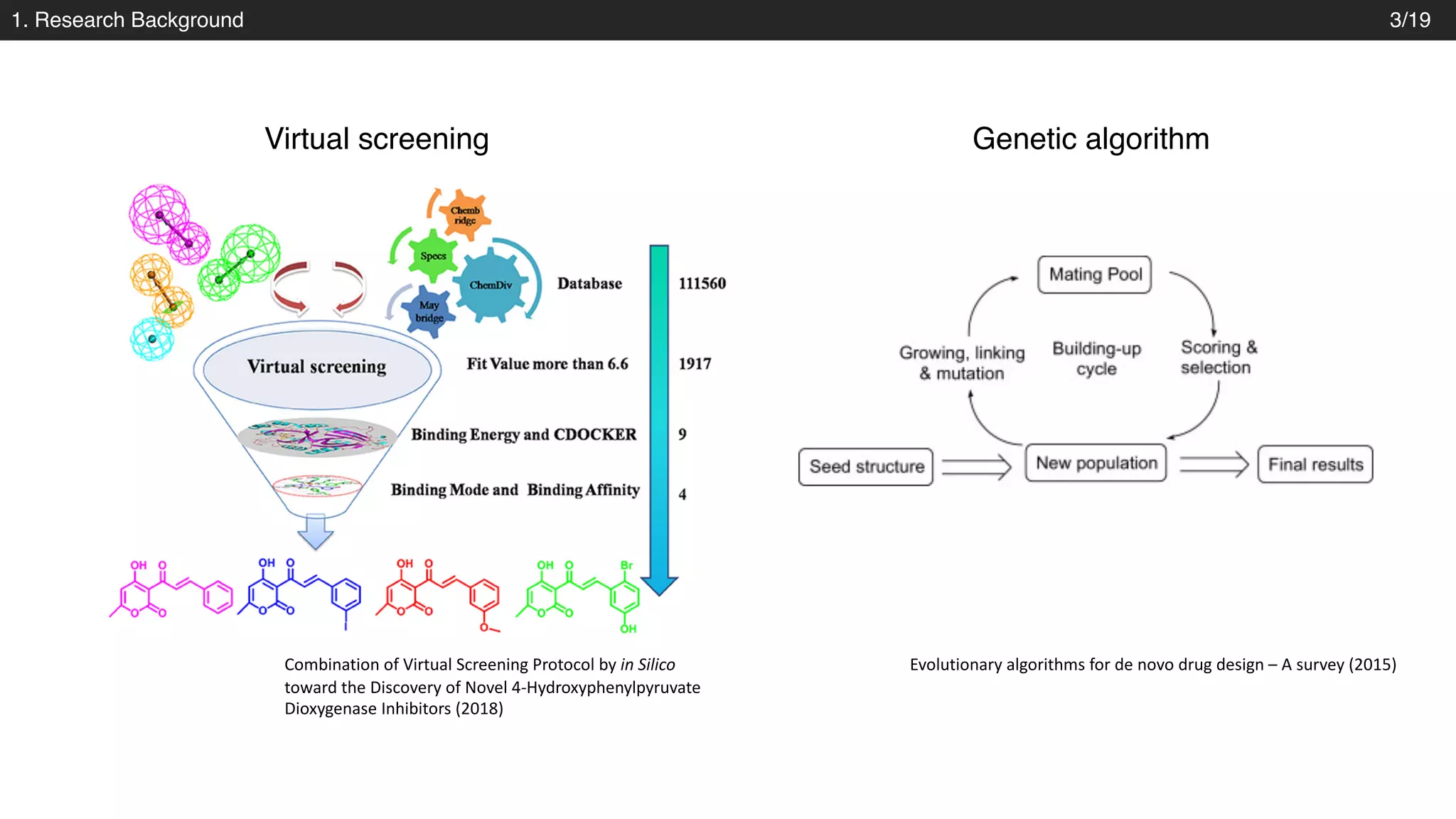

![2. Methods
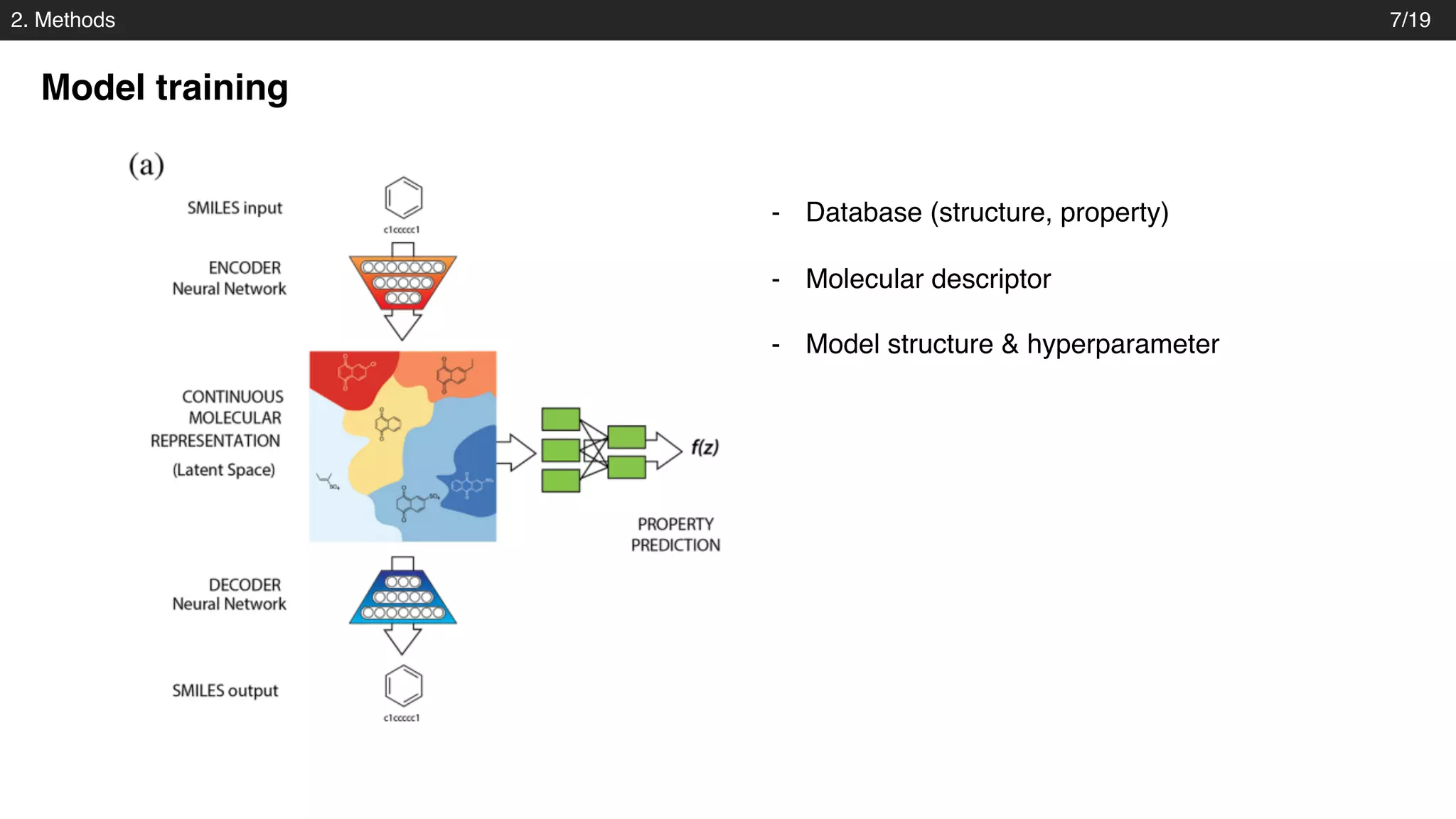
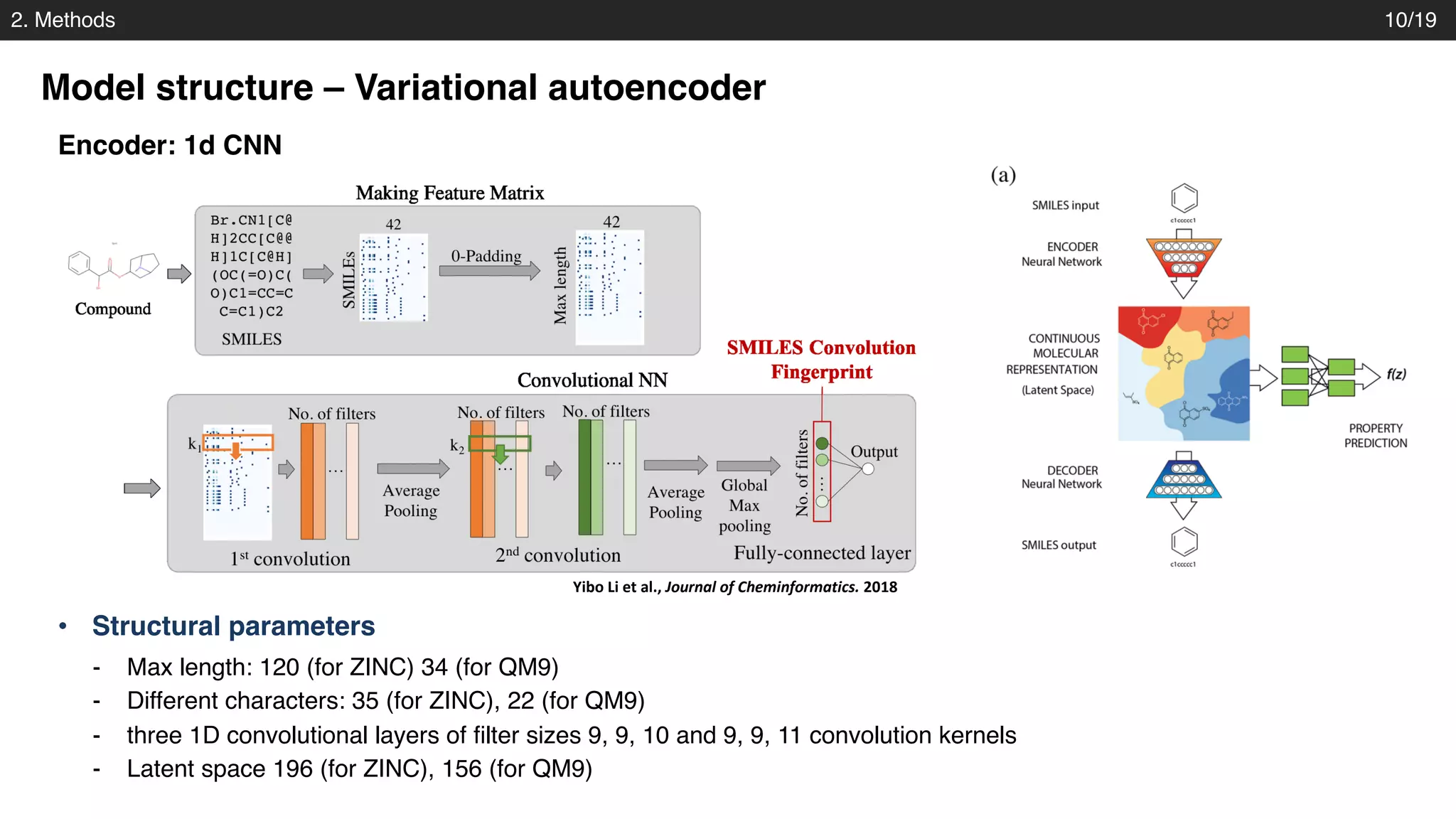
Model structure – Variational autoencoder
11/19
Decoder: gated recurrent unit (GRU)
Josep Arús-Pous et al., Journal of Cheminformatics. 2018
- into three layers of gated recurrent unit (GRU) networks with hidden dimension of 488.
- Property prediction : fully connected layers [1000, 1000]
• Structural parameters](https://image.slidesharecdn.com/pr173automatic-chemical-designjoo-190623162815/75/PR173-Automatic-Chemical-Design-Using-a-Data-Driven-Continuous-Representation-of-Molecules-13-2048.jpg)