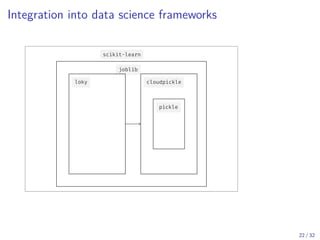
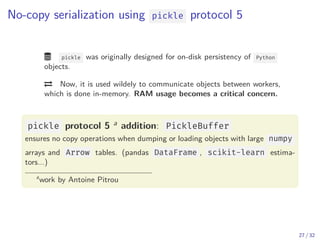
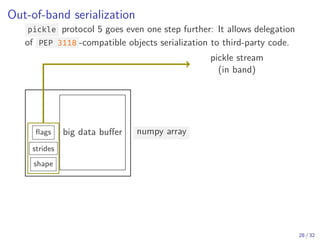
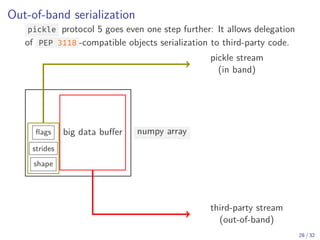
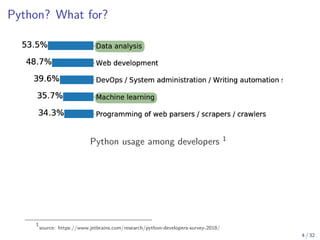
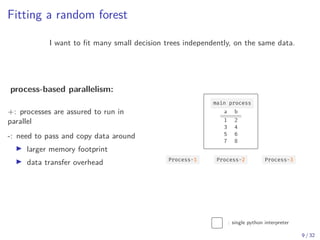
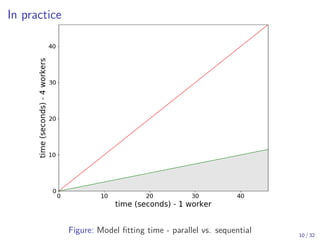
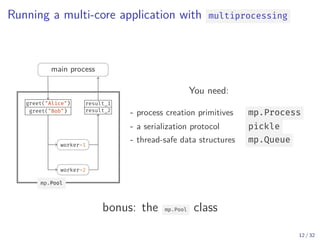
The document discusses parallel computing in Python. It provides an overview of parallelization libraries in Python like multiprocessing and concurrent.futures. It describes different approaches to parallelization using threads versus processes. Thread-based parallelism allows sharing memory but the Global Interpreter Lock limits parallel execution, while process-based parallelism ensures true parallel execution but requires data copying between processes. It also discusses challenges like serialization and portability across environments. Alternatives like loky provide a more robust process pool executor for data science tasks.


















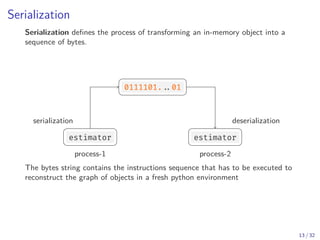
![The pickle protocol
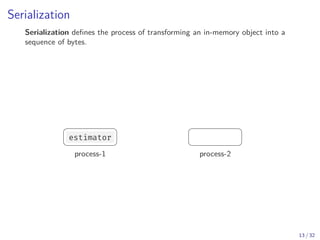
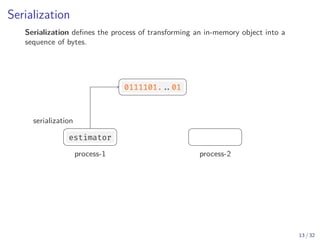
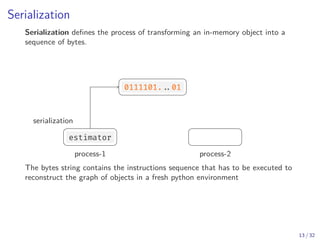
Python defines a serialization protocol called pickle , and provides an
implementation of it in the standard library.
import pickle
s = pickle.dumps([1, 2, 3]) # serialization (pickling) step
s
b'x80x03]qx00(Kx01Kx02Kx03e.'
depickled_list = pickle.loads(b'x80x03]qx00(Kx01Kx02Kx03e.')
depickled_list
[1, 2, 3]
14 / 32](https://image.slidesharecdn.com/main-190910133400/85/Parallel-computing-in-Python-Current-state-and-recent-advances-33-320.jpg)