The document provides an overview of nucleic acid metabolism and the synthesis of purines and pyrimidines. It discusses how nucleotides serve as building blocks for nucleic acids and how they are synthesized through both de novo and salvage pathways. The key steps in purine synthesis include the production of IMP from PRPP and glutamine, followed by conversion to AMP and GMP. Purines can also be salvaged from nucleic acid breakdown. Deoxyribonucleotides are synthesized from ribonucleotides by the enzyme ribonucleotide reductase. Defects in nucleotide synthesis can cause diseases.
















![2. Synthesis of 5-phosphoribosylamine
Synthesis of 5-phosphoribosylamine from PRPP and glutamine.
The amide group of glutamine replaces the pyrophosphate
group attached to carbon 1 of PRPP. This is the committed step in
purine nucleotide biosynthesis.
The enzyme, glu-tamine:phosphoribosylpyrophosphate
amidotransferase, is inhibited by the purine 5 -nucleotides AMP
and [GMP], the end products of the pathway.
The rate of the reaction is also controlled by the intracellular
concentration of PRPP. Any small change in the PRPP
concentration causes a proportional change in rate of the
reaction (acc. To Michaelis Menton eq.)
18
Amidophosphoribosyl
Transferase](https://image.slidesharecdn.com/nucleicacidmetabolism-210615154859-220426173137/75/Nucleic-Acid-Metabolism-18-2048.jpg)
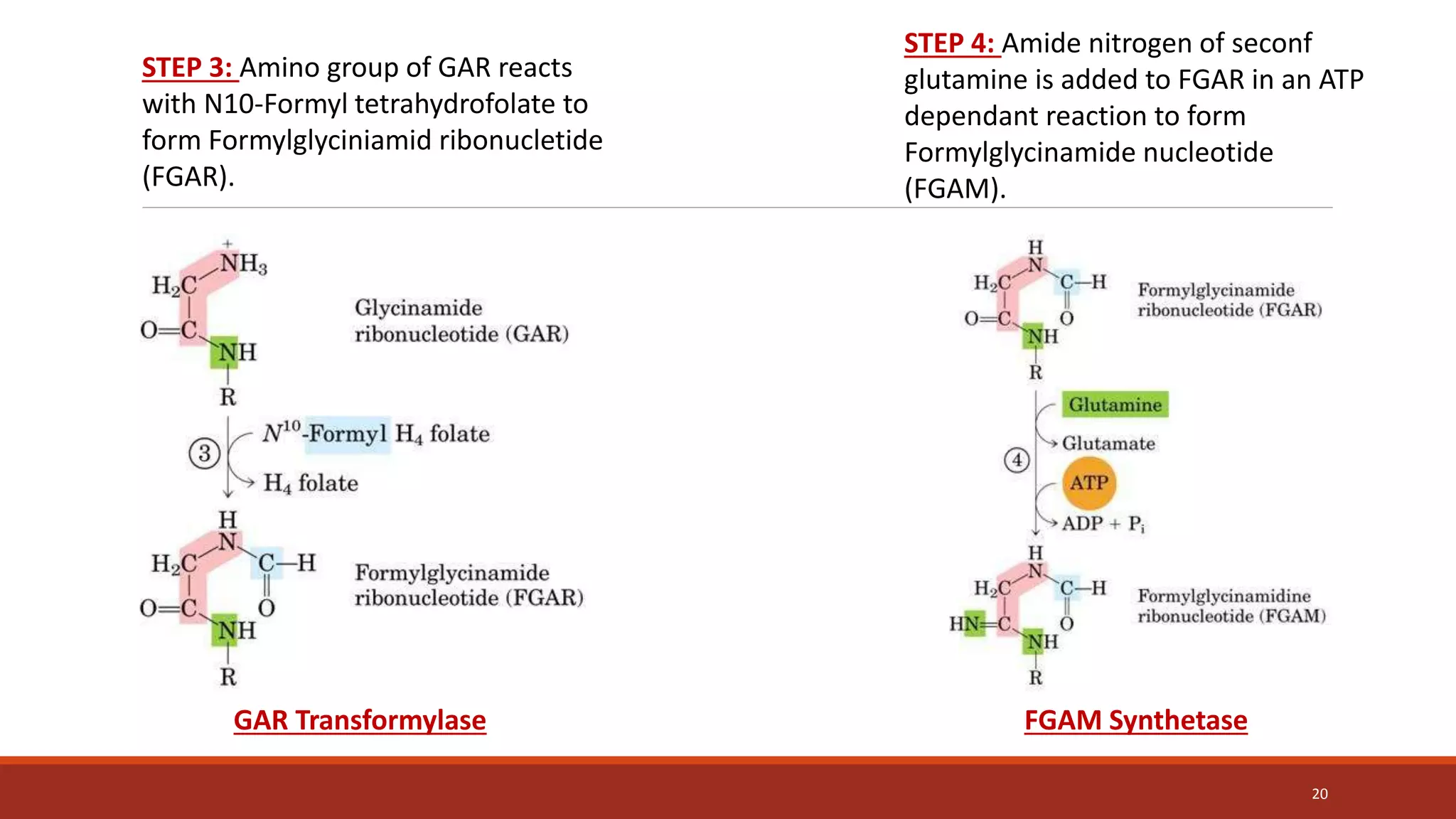
![Synthesis of IMP, the “parent” purine nucleotide
The next nine steps in purine nucleotide biosynthesis leading to the synthesis of
inosine monophosphate ([IMP] whose base is hypoxanthine).
Four steps in this pathway require ATP as an energy source, and two steps in the
pathway require N10-formyltetrahydrofolate as a one-carbon donor. (Hypoxanthine is
found in tRNA).
The next 9 steps out of 11 are elaborated stepwise in next slides.
19](https://image.slidesharecdn.com/nucleicacidmetabolism-210615154859-220426173137/75/Nucleic-Acid-Metabolism-19-2048.jpg)







![1. Salvage of purine bases to nucleotides
Two enzymes are involved: adenine phosphoribosyltransferase (APRT) and
hypoxanthine-guanine phosphoribosyltransferase (HGPRT). Both enzymes use
PRPP as the source of the ribose 5-phosphate group.
The release of pyrophosphate and its subsequent hydrolysis by
pyrophosphatase makes these reactions irreversible. [Note: Adenosine is the
only purine nucleoside to be salvaged. It is phosphorylated to AMP by adenosine
kinase.]
30](https://image.slidesharecdn.com/nucleicacidmetabolism-210615154859-220426173137/75/Nucleic-Acid-Metabolism-30-2048.jpg)