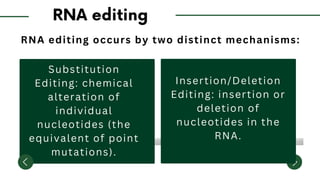
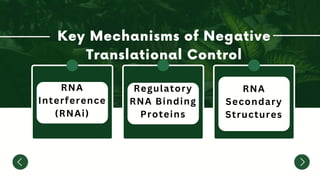
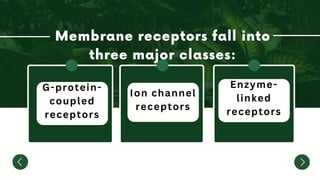
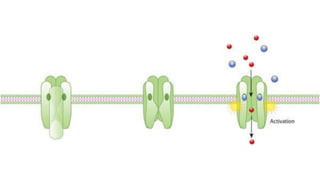
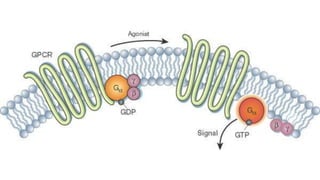
The document discusses the mechanisms of transcriptional control, RNA splicing, and gene expression regulation in cellular biology. It outlines how transcription factors, chromatin remodeling, and RNA editing influence gene activity, while also detailing processes such as negative translational control through RNA interference. Additionally, the document covers various types of cell signaling methodologies and the role of receptors in responding to stimuli.