This study used mass spectrometry to validate genome annotations of nine diverse mycobacteriophages and identify their expressed proteins during infection of Mycobacterium smegmatis. Peptides matching both characterized and uncharacterized putative phage proteins were detected, validating the genome annotations. Over 100 peptides were identified for some phages, including proteins for virion structure, DNA replication, and host lysis. The study demonstrates mass spectrometry can rapidly validate phage genome annotations and identify expressed proteins, aiding characterization of phage-host interactions.




![- 5 -
Background
Bacteriophages or phages are the viruses that infect bacteria. Similar to other viruses,
a phage is constructed of two components: a genome of nucleotide material and a
capsid made of proteins [1]. The phage genome can be either single- or double-
stranded DNA or RNA and is dependent upon the host for replication and the
production of new phage particles. Interactions between the host and the phage result
in either lytic or lysogenic phage life cycles. In a lytic cycle, the phage genome is
replicated, transcribed, and translated using the bacterial machinery. Phage particles
are then assembled and released out of the bacterium. In contrast, during the lysogenic
cycle, the phage genome is integrated into the bacterial chromosome. Under certain
conditions, the integrated phage genome can be released from the host chromosome
and subsequently enter a lytic cycle [2].
Mycobacteriophages, phages that can infect Mycobacteria, have been studied
extensively. Novel phages are isolated from environmental samples, and phage
genomes are sequenced and annotated [3]. Isolated mycobacteriophages can be
categorized into clusters through a four-step analysis at the nucleotide level: dot-plot
comparison of all genomes with one another, pairwise average nucleotide identities
(ANI), pairwise genome map comparisons, and gene content analysis [3]. Functions
of annotated gene products are predicted through bioinformatics analysis and
homologous proteins are assembled into different phamilies [4]. Wet lab
experimentation is still needed to validate the genome annotations and further
characterize the putative phage proteins and their function. Previous studies employed
mass spectrometry (MS) to examine phage proteins [5], however, since analysis
focused solely on purified phage particles, only virion and structural proteins were
examined. Exploration of the remaining phage proteins is challenging since protein](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-5-2048.jpg)
![- 6 -
production depends upon complex interactions between the phage and its bacterial
host. Thus the availability and concentration of the proteins in phage lysates is
unpredictable. In this research, proteins were extracted from the phage-bacteria
mixture during phage infection, and further analyzed by mass spectrometry.
Results
Sample Preparation
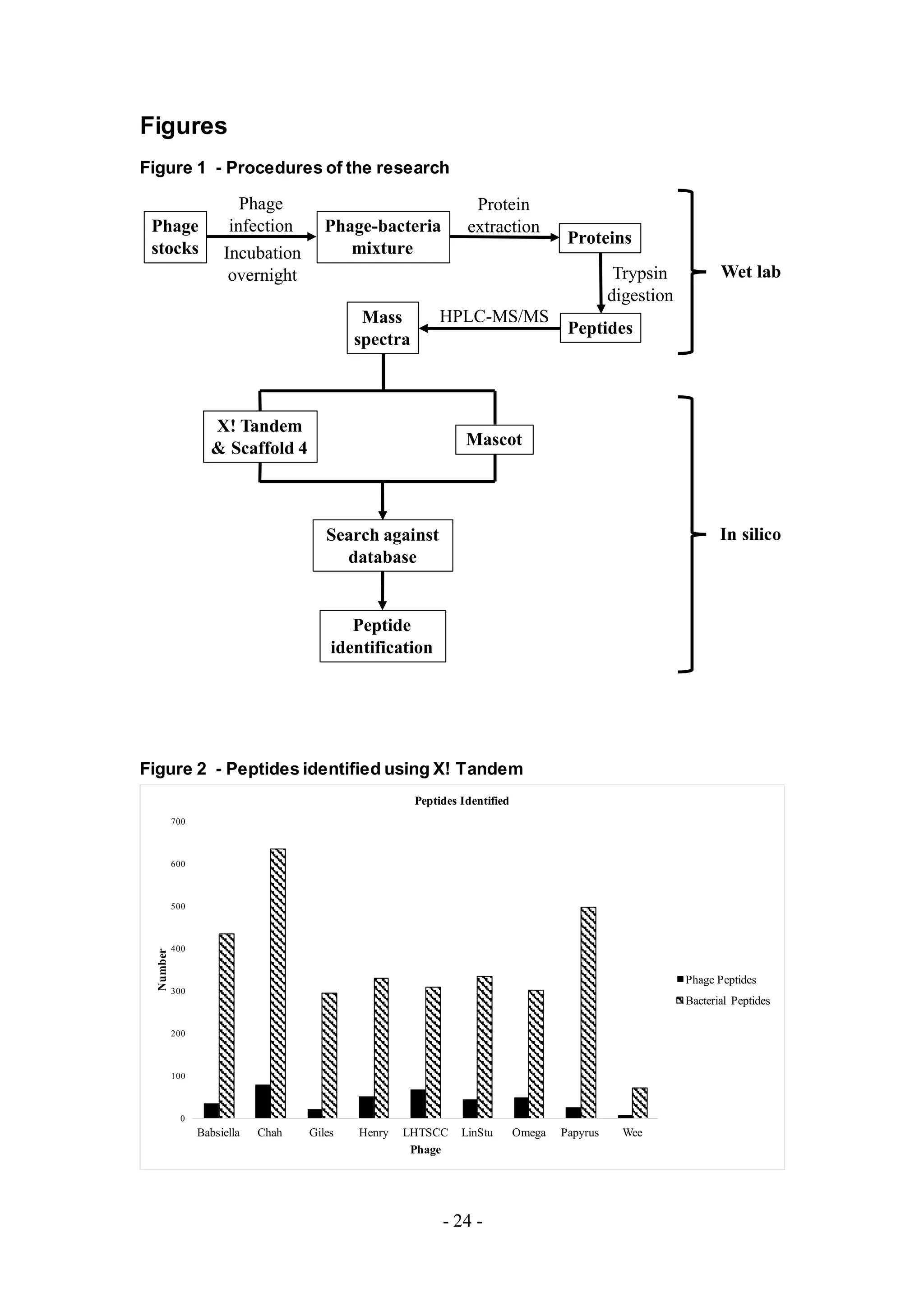
The overall experimental process is shown in Figure 1. Phages from diverse clusters
(Table A1) were obtained from Prof. Graham Hatfull’s research group through the
HHMI SEA-PHAGES program. Individual phage stocks were incubated separately
with M. smegmatis cell culture and provided maximum time for infection and
production of phage proteins by the infected bacteria. Non-infected M. smegmatis cell
culture was used as a negative control. Total proteins of the phage-bacteria mixture
were extracted using high pressure to break the cell membrane and optimize the yield
of proteins, digested using trypsin and loaded to mass spectrometer for analysis.
Figure 1. Experimental Process
The MS method was designed and performed at Purdue University [6]. Data analysis
using X! Tandem was adapted from Pope, et al [7].
Identification of phage peptides by Mass Spectrometry
To identify the peptides that were expressed in the phage-bacteria mixture, the
software X! Tandem was used to search the mass spectra raw data against a database
containing the six-frame translation of the sample phage genomes, the annotated
proteins of M. smegmatis, and the proteins from the comprehensive protein database,](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-6-2048.jpg)
![- 7 -
UniProt, as published previously [7]. In the output of X! Tandem, peptides from both
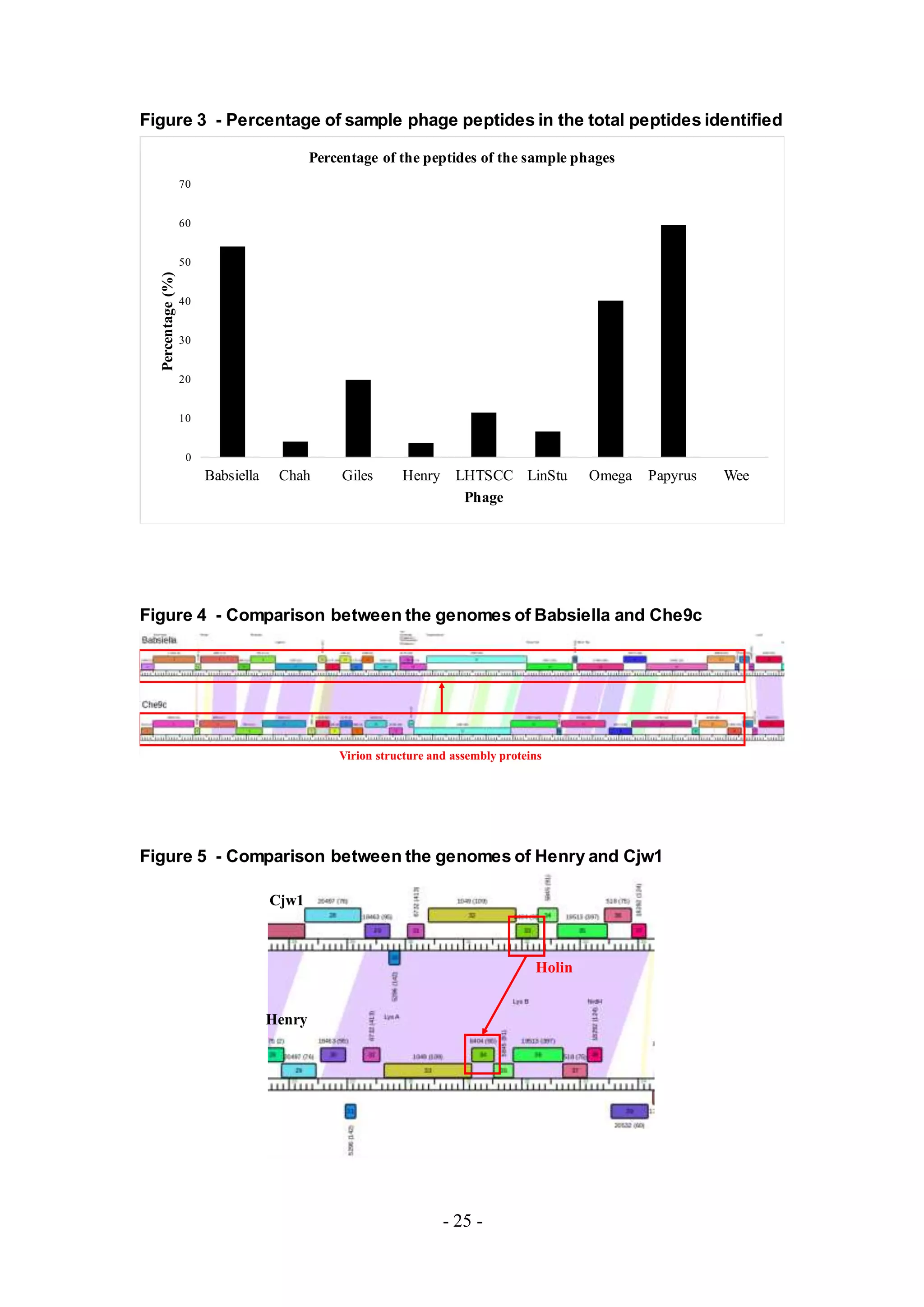
the corresponding phages and the host mycobacteria were detected. The number of
peptides, however, from the host was much larger than the amount of detected
peptides from the phage (Figure 2). The total number of identified phage peptides in
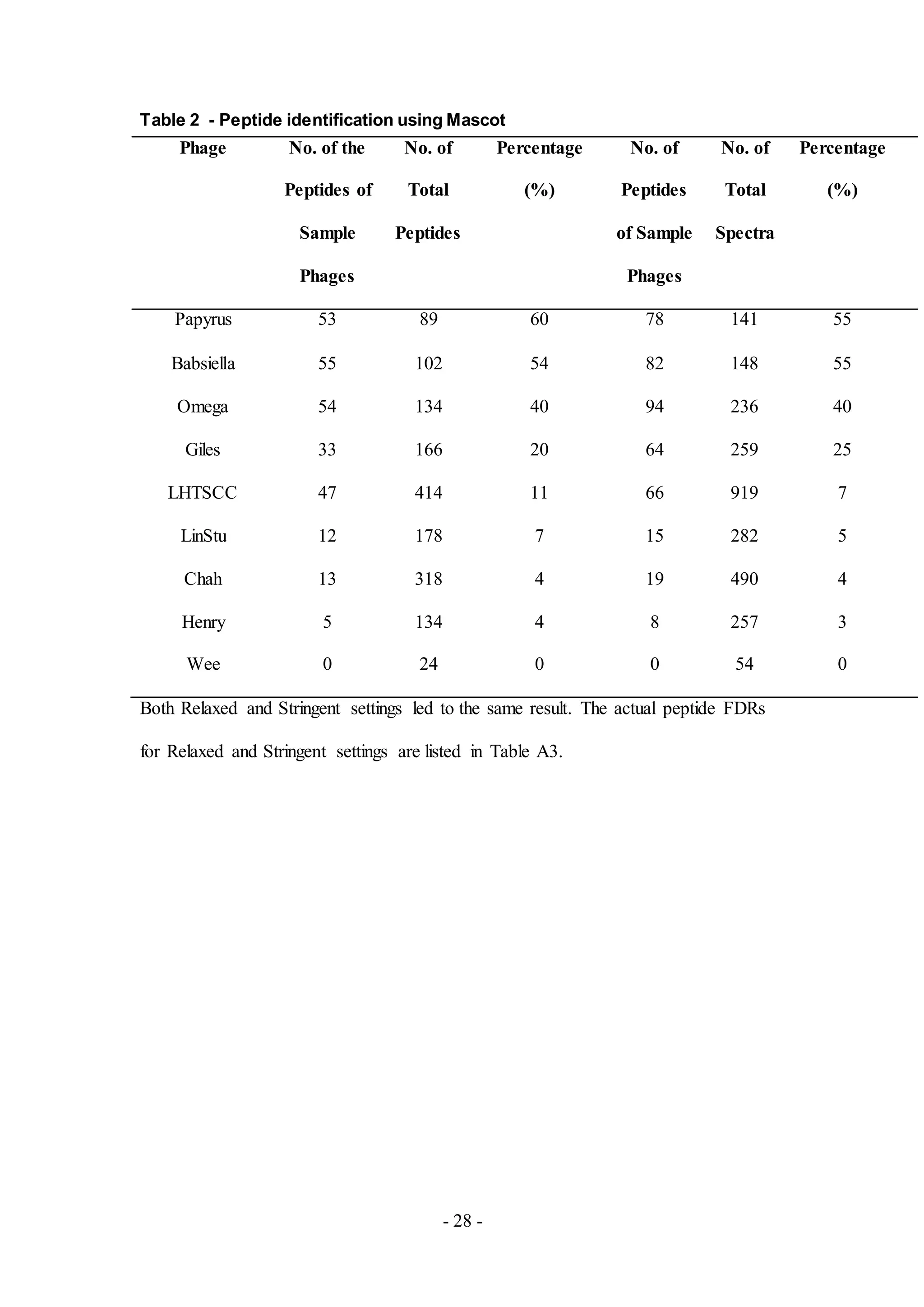
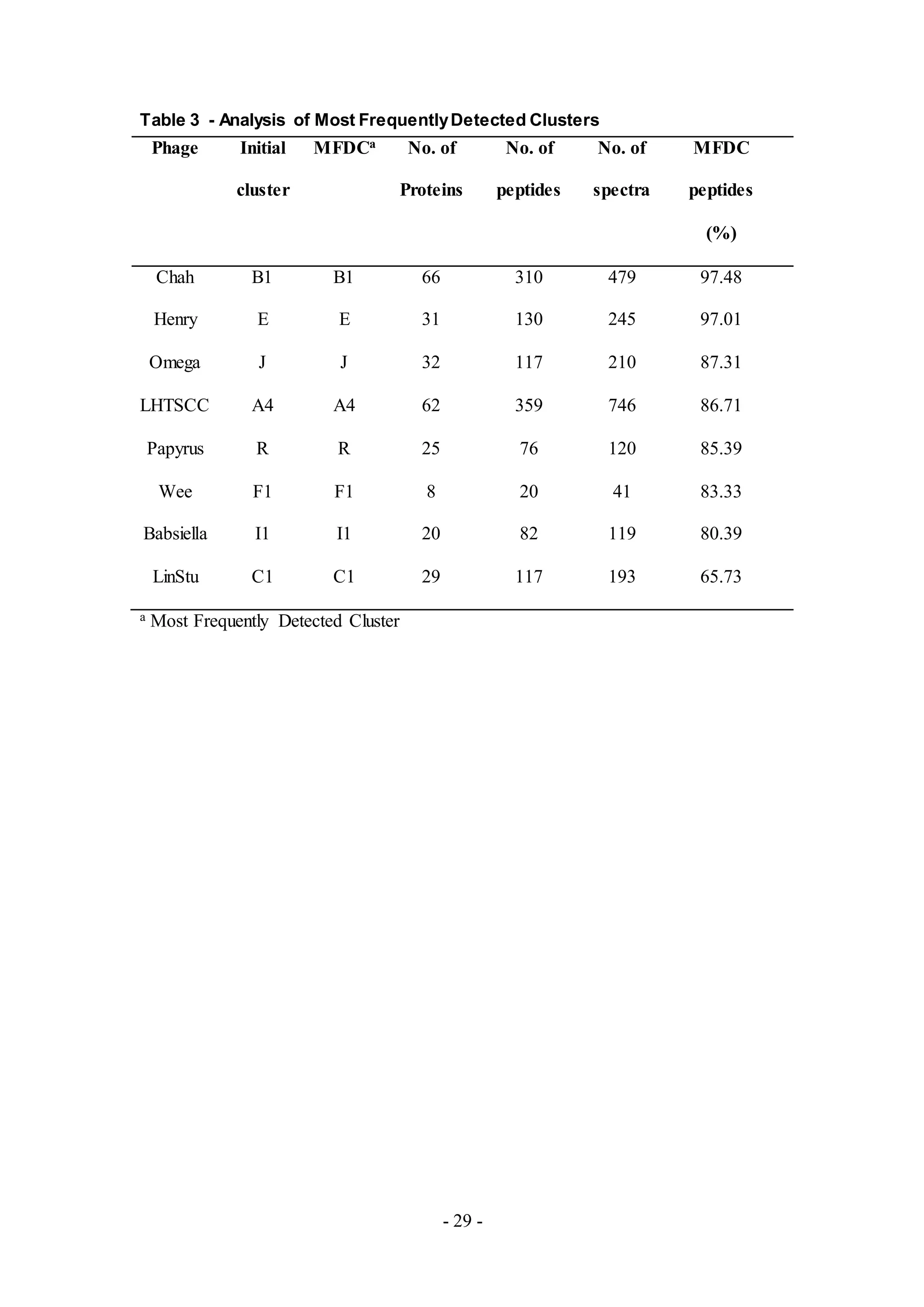
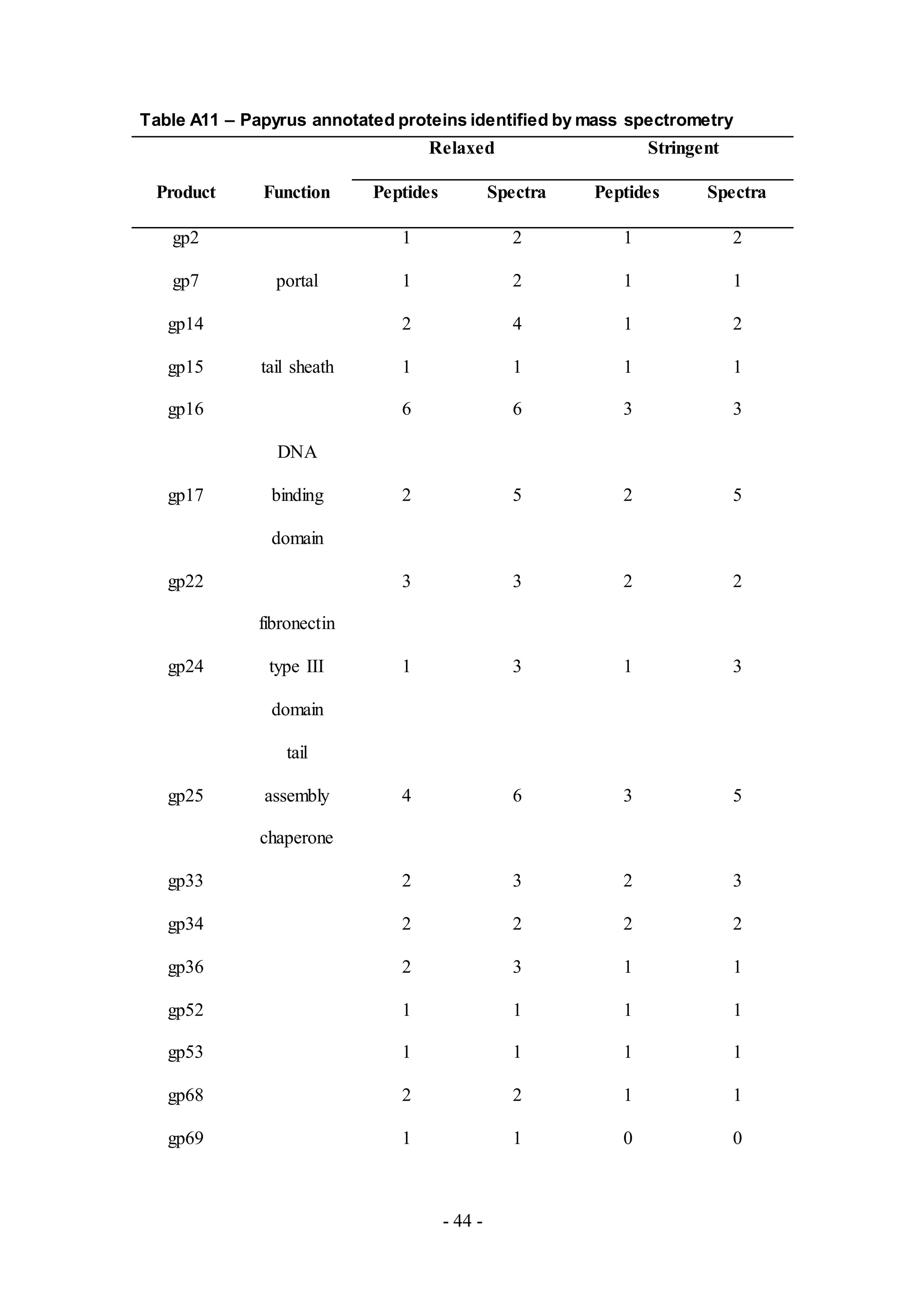
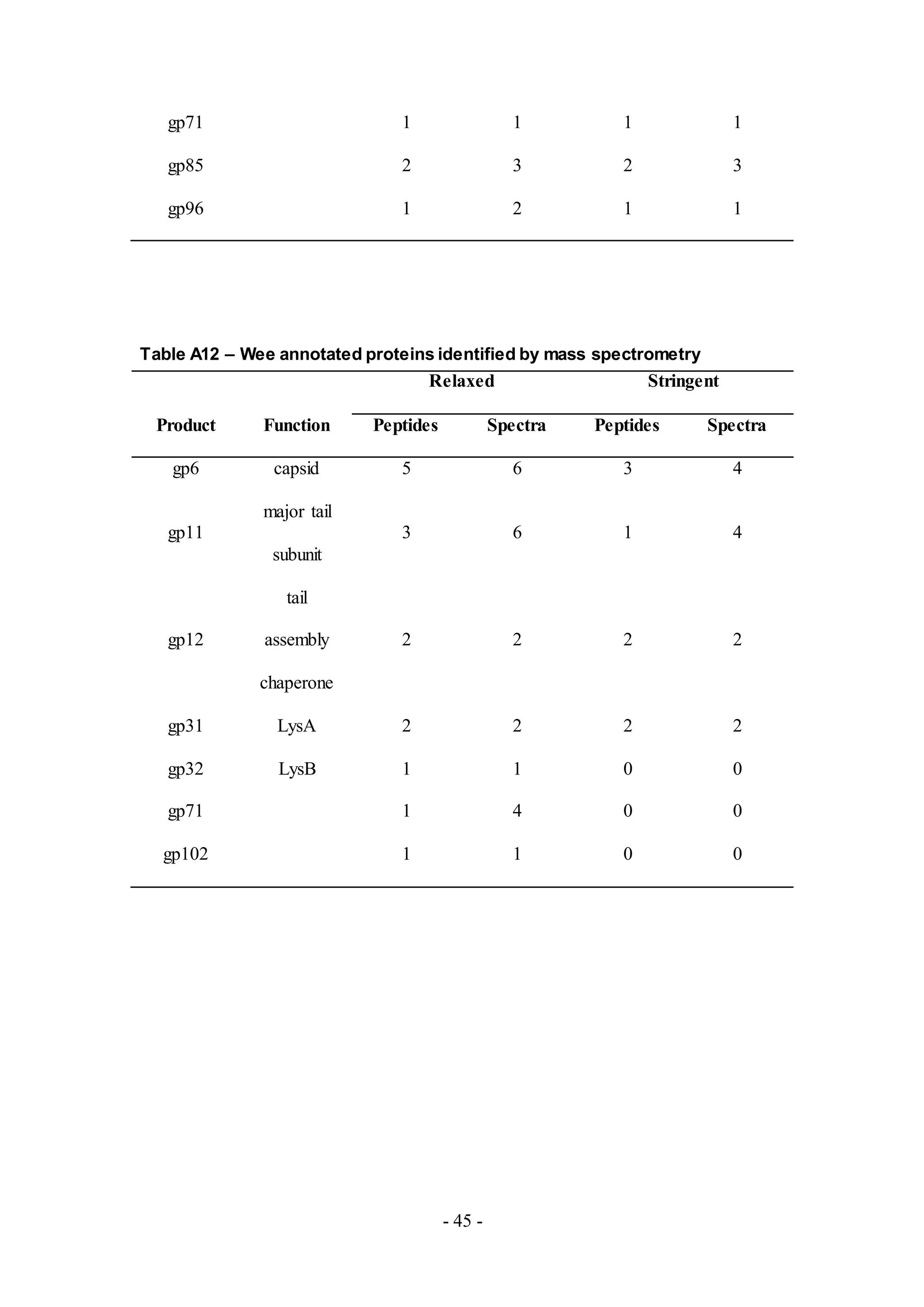
all samples is listed in Table 1.
Figure 2. Peptides identified by using X! Tandem.
The number of identified phage peptides was low in comparison to the number of
identified bacterial peptides. Additional software, Scaffold 4, was used for further
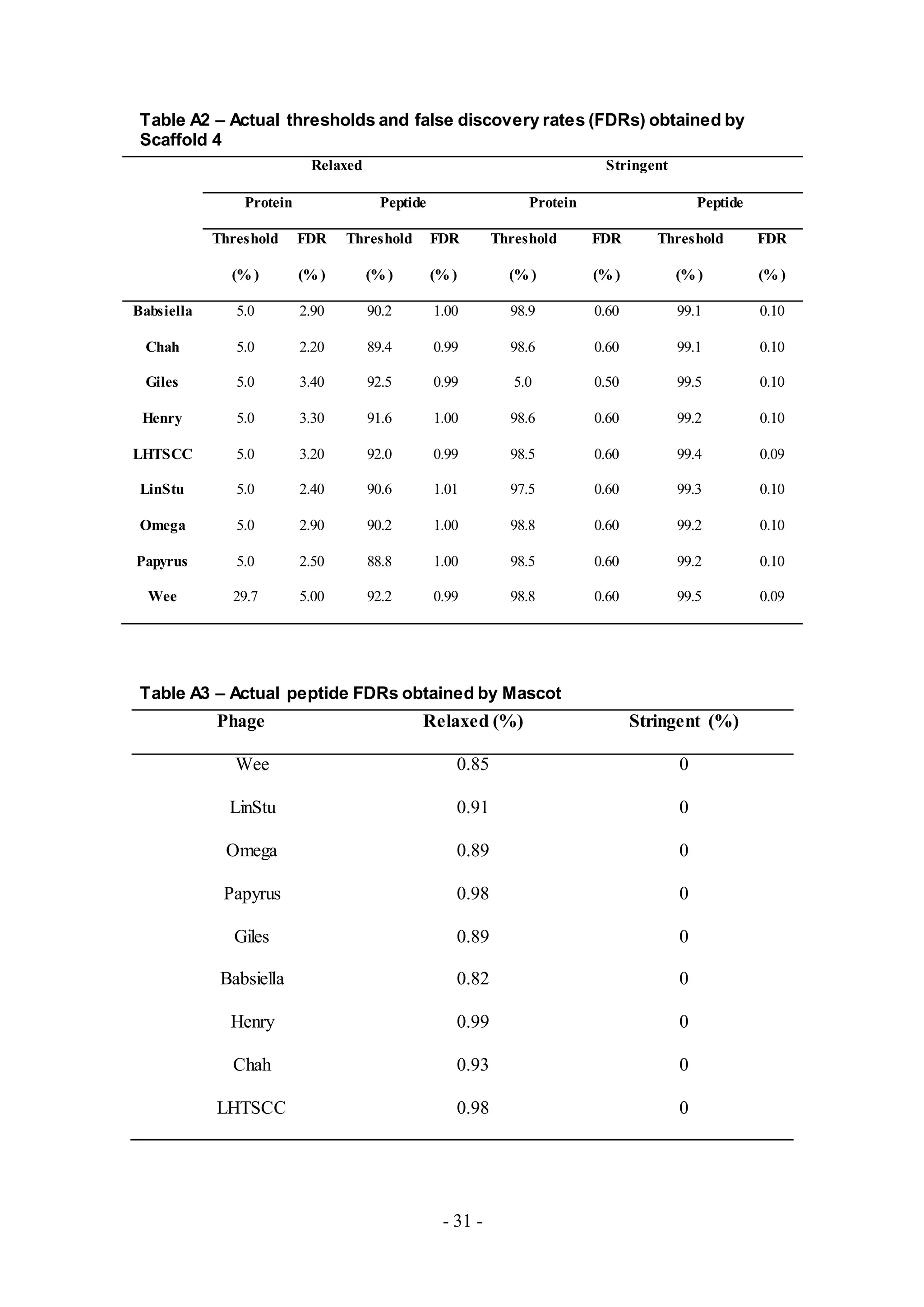
analysis and adjustment of False Discovery Rates (FDRs) and the results obtained
using the Stringent setting is shown. All phage proteins identified based upon the
peptides detected from MS are listed in Tables A4-A12.
Identification of phage proteins
The phage peptides identified with X! Tandem were labeled in the genome map of the
phages to compare the expressed phage proteins with the predicted proteins from the
phage genome annotation. Four proteins were identified from phage Wee, more than
ten proteins were detected in Babsiella, Giles, Henry, LHTSCC, LinStu, Omega and
Papyrus, and more than thirty proteins were observed in Chah. Although only a
portion of the putative proteins predicted from the genome annotations were
identified, the detected peptides validated the existing genome annotations for the
corresponding genes. As expected, putative virion structure and assembly proteins
were detected in all samples, due to the presence and production of phage virions.
Non-structural proteins predicted by the genome annotations were also detected as
described in the following paragraphs.](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-7-2048.jpg)
![- 8 -
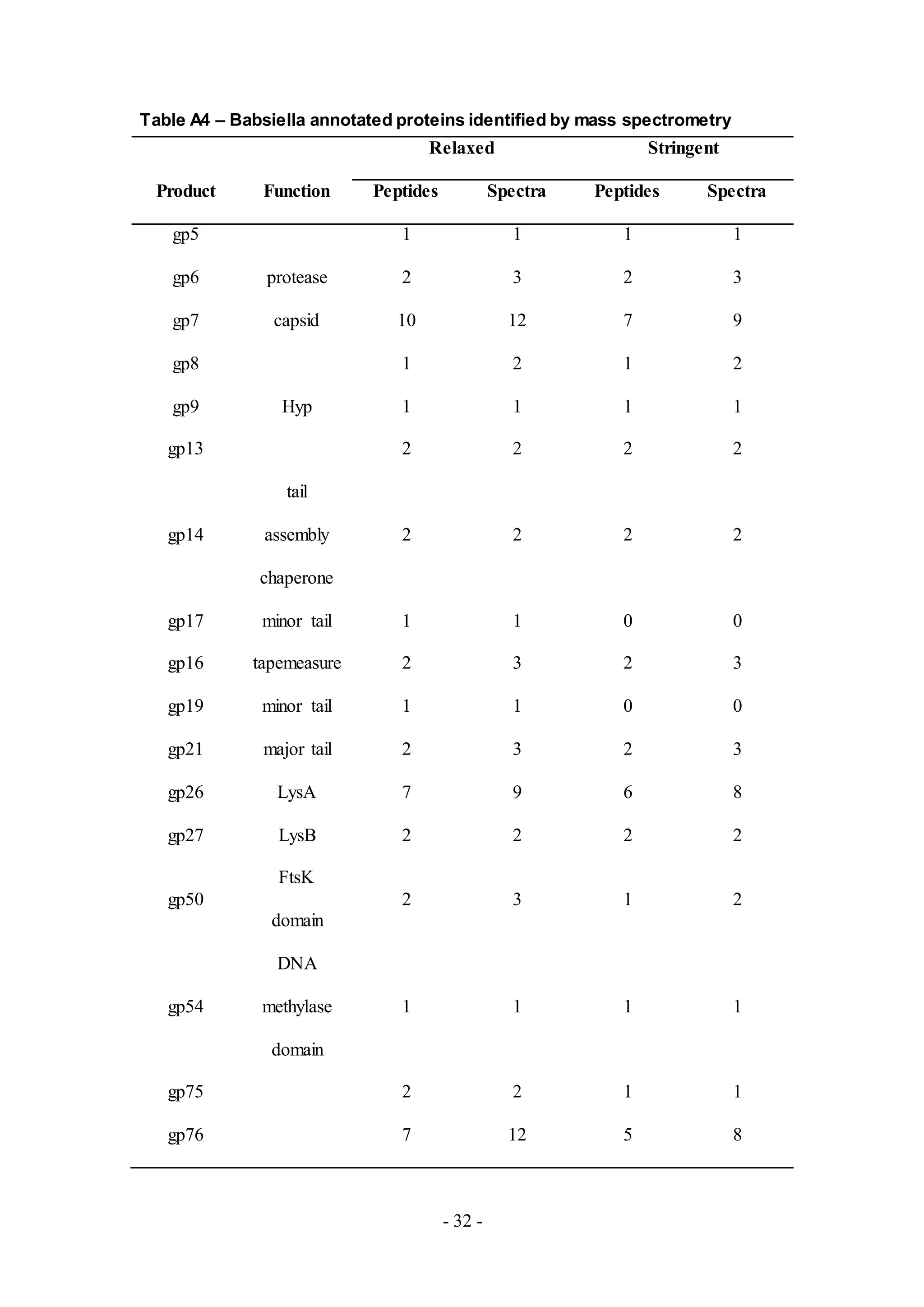
Fifteen proteins were detected in Babsiella (Table A4), including LysA and LysB,
proteins that prompt host cell lysis. Other proteins were putative virion structure and
assembly genes based upon homology with another phage member of Cluster I,
Che9c. The first twenty two genes from Che9c are virion structure and assembly
proteins [4] and based upon a comparison to Babsiella using Phamerator, the putative
proteins have a similar structure and distribution and belong to conserved phamilies
(Figure 4). Therefore, although gp5 (gene product of gene 5), gp8, and gp13 of
Babsiella are functionally uncharacterized, their putative proteins may be associated
with virion structure and assembly based upon the comparison in Phamerator. The
remaining proteins identified, gp75 and gp76, have unknown functions.
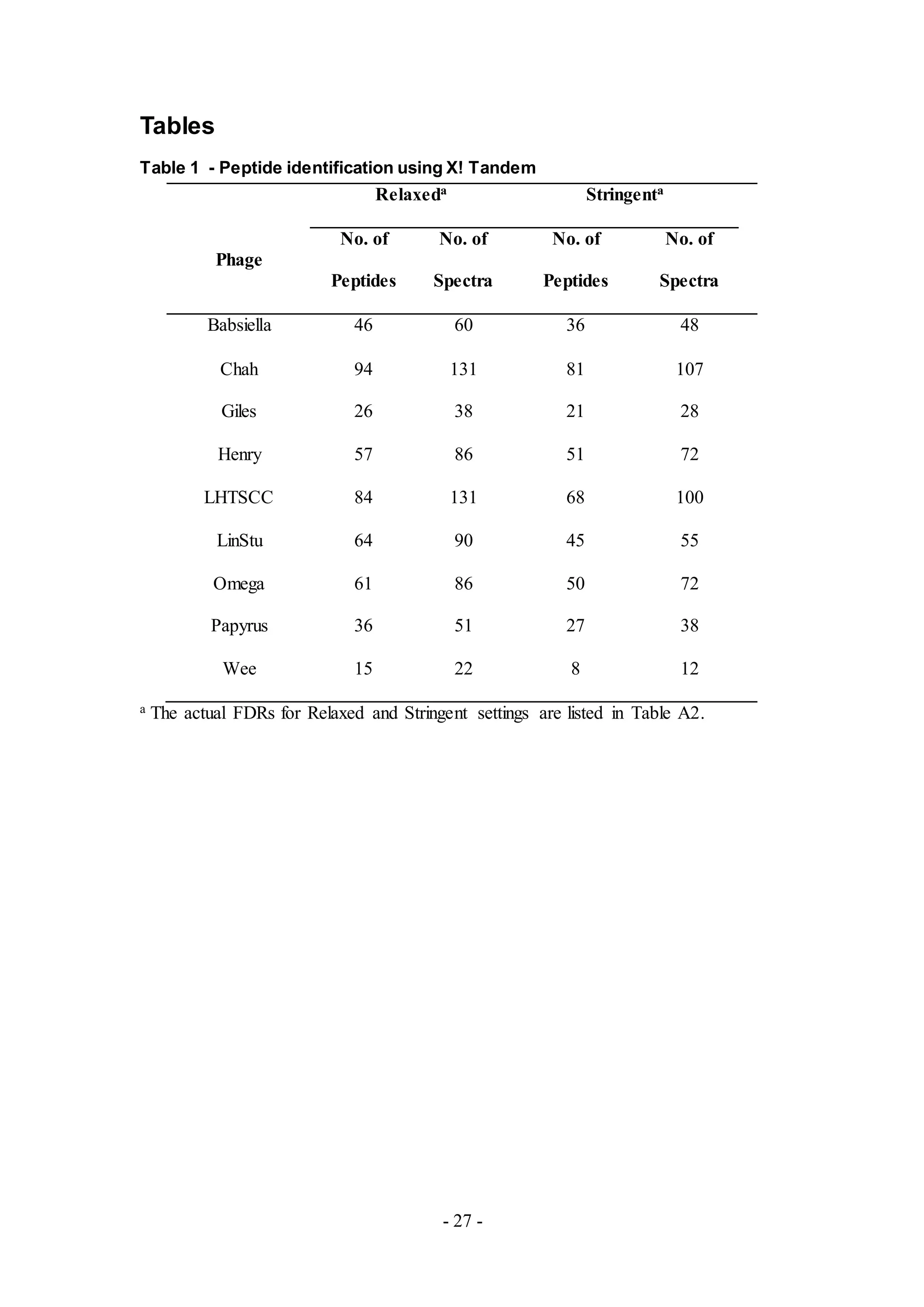
Figure 4. Comparison between the genomes of Babsiella and Che9c
The genomes of Babsiella and Che9c were compared using Phamerator [9]. Most of
the first 22 genes in both genomes belong to the same phamilies (same color and
phamily number) and have similar distribution.
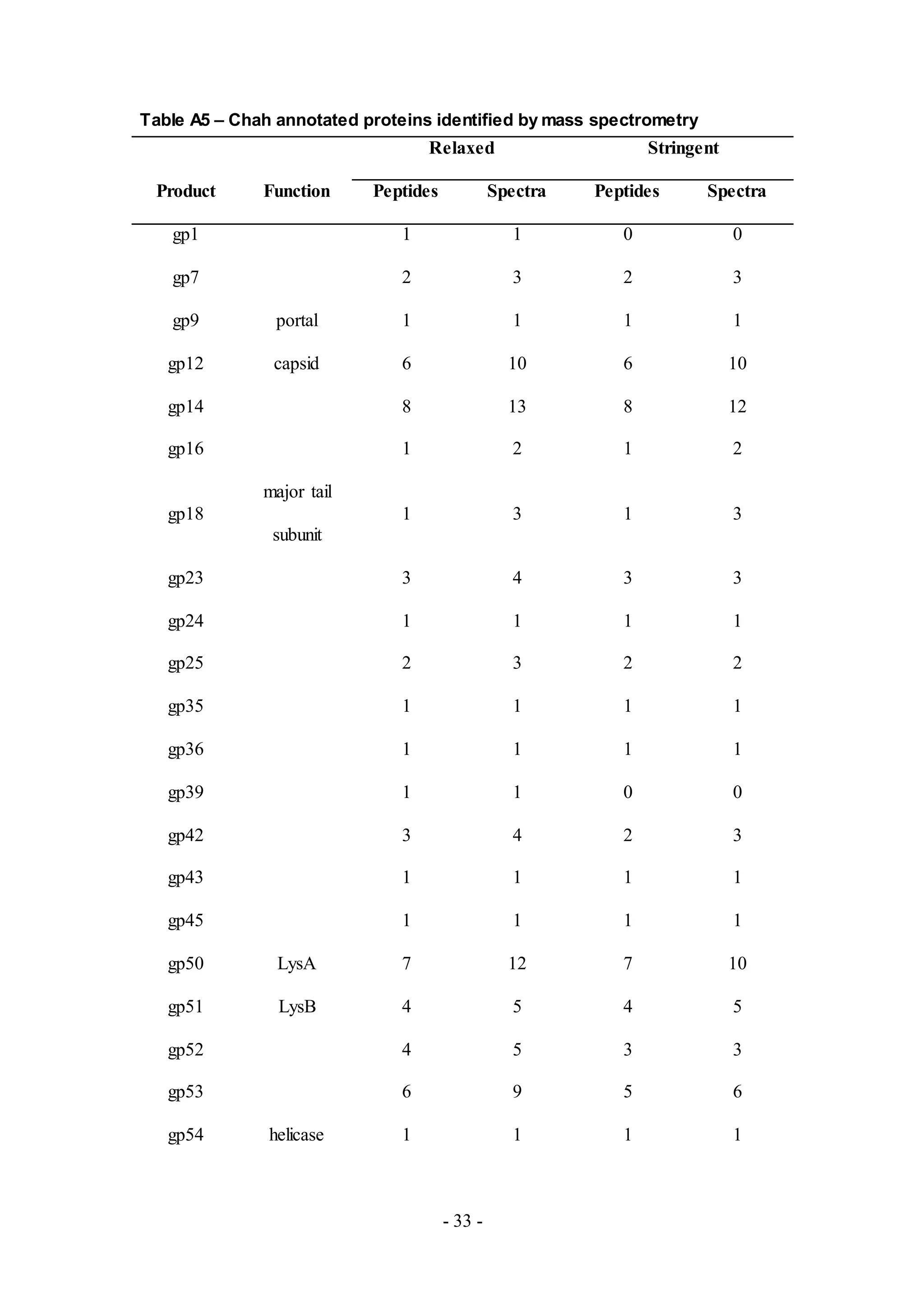
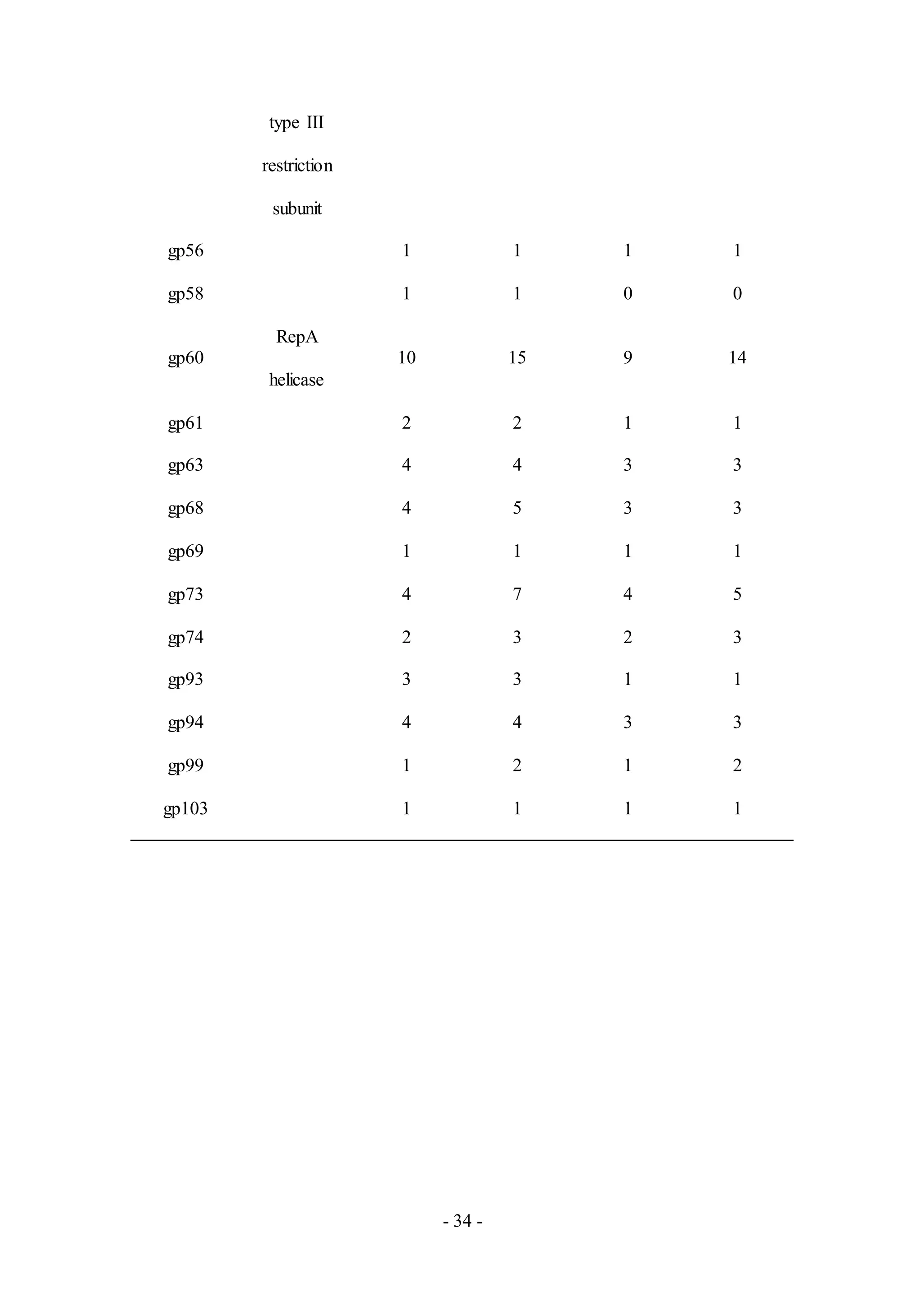
Thirty one proteins were detected from Chah (Table A5), a phage from Subcluster B1.
Among the detected proteins, only seven had putative functions assigned based on
genome annotation efforts. Potential functions include proteins responsible for: virion
structure and assembly such as portal (gp9), capsid (gp12) and major tail subunit
(gp18); host cell lysis such as LysA (gp50) and LysB (gp51); and DNA replication
such as helicase type III restriction subunit (gp54) and RepA helicase (gp60).
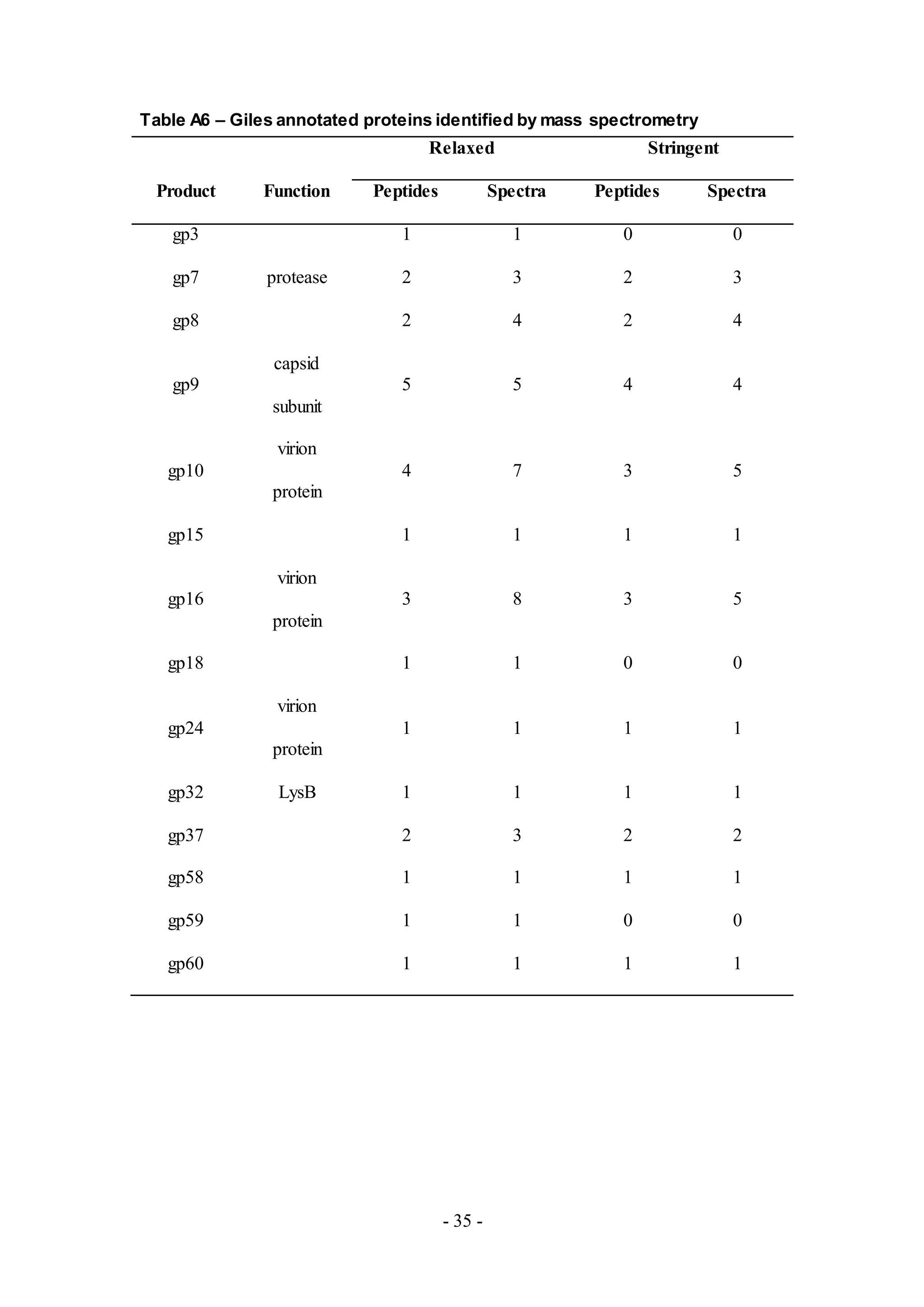
Eleven proteins were detected from Giles (Table A6), one of five phages from Cluster
Q. Seven of the eleven proteins detected are putative virion structure and assembly](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-8-2048.jpg)
![- 9 -
proteins. In addition, Clp protease (gp7), a protein involved in a protease-required
capsid assembly process [4], and LysB (gp32) were also detected along with
functionally uncharacterized proteins, gp37, gp58, and gp60.
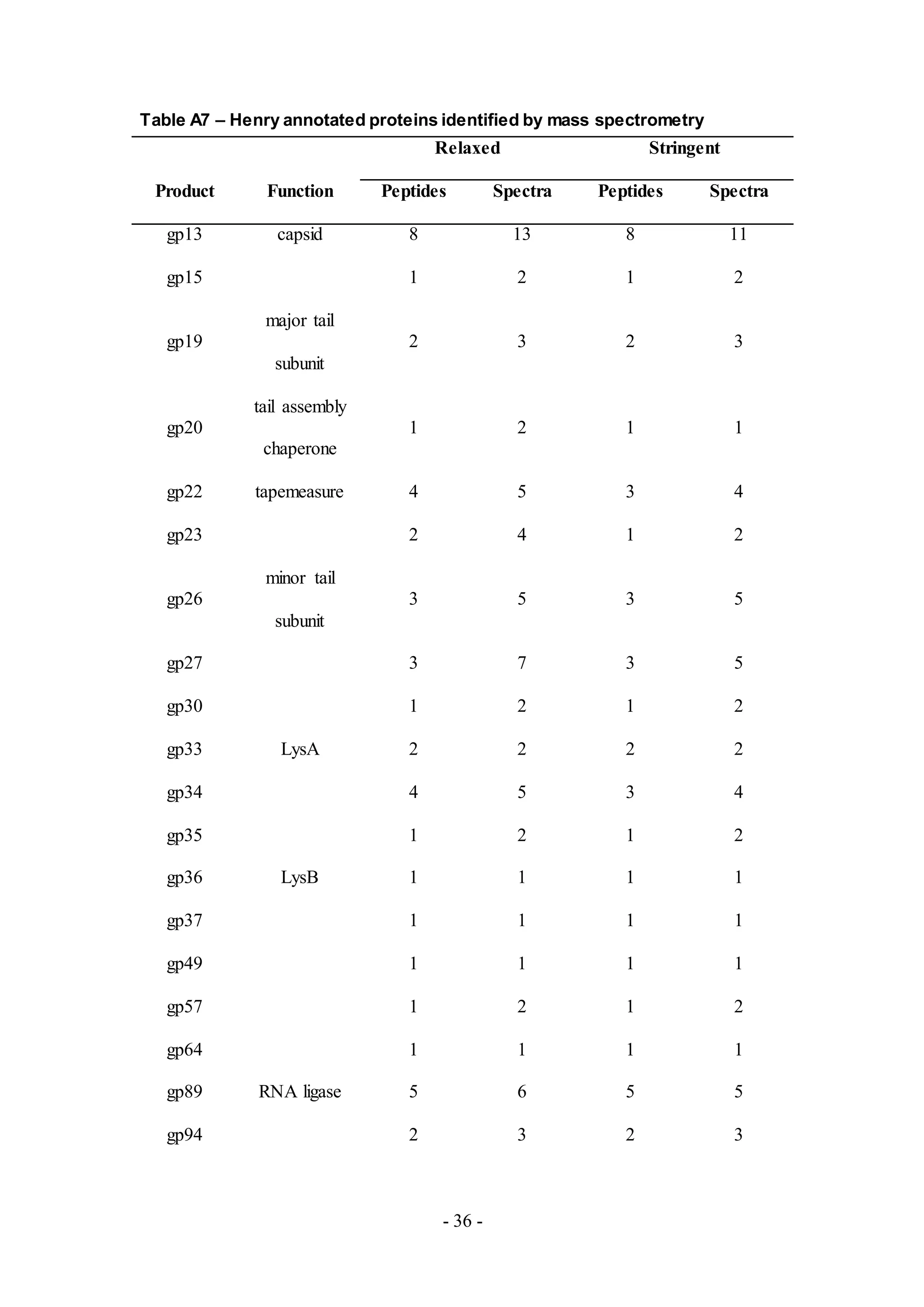
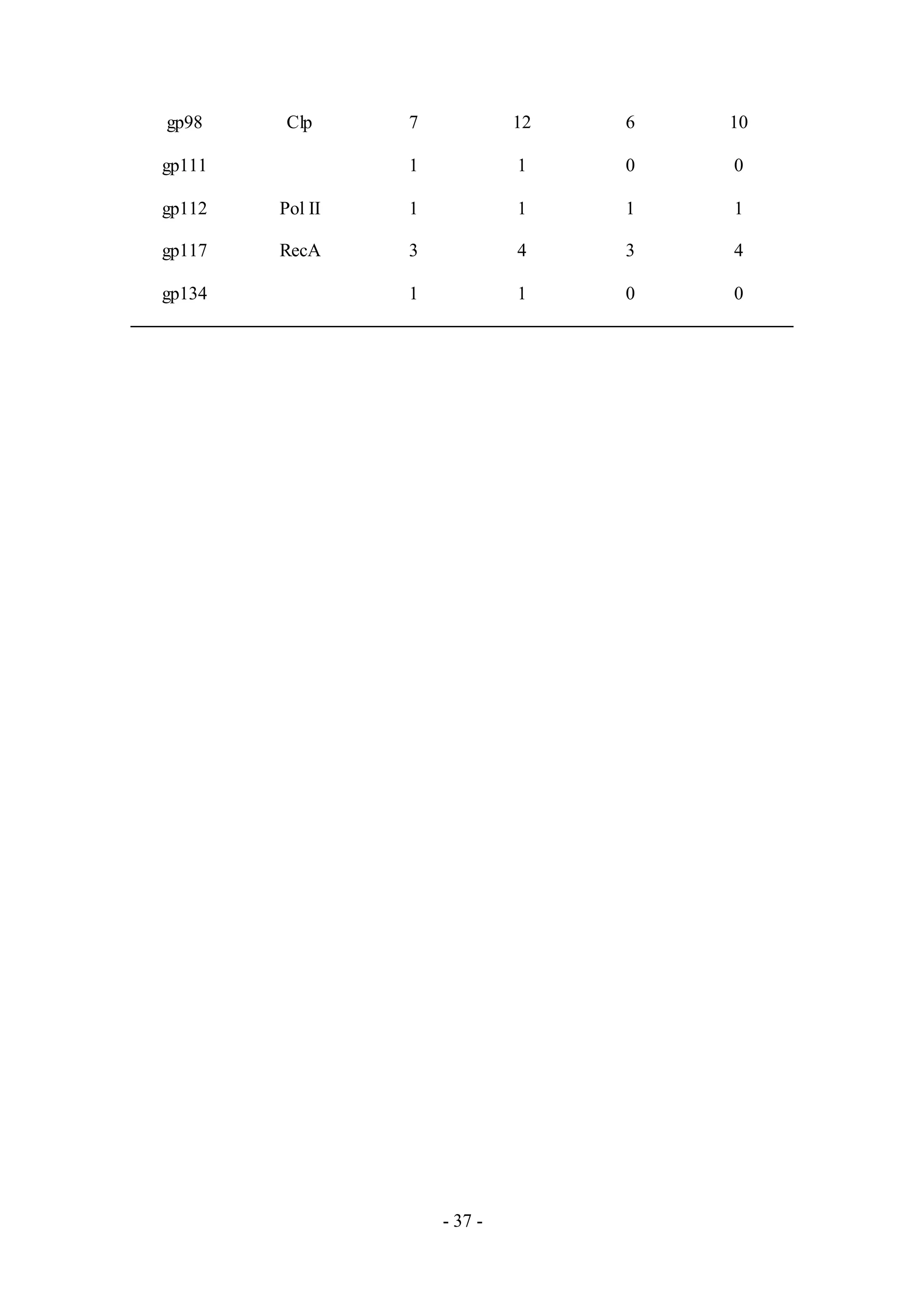
Twenty two proteins were detected from the phage sample, Henry (Table A7), a
member of Cluster E. Putative proteins for virion structure and assembly, lysis
cassette and DNA replication were detected, similar to results from the phage sample,
Chah. Gp34 was also detected but it is not functionally characterized based upon the
genome annotation. Performing a genome comparative analysis with Phamerator,
however, suggests that gp34 from Henry is a holin, based upon homology with the
holin (gp33) of another phage from Cluster E, Cjw1 (Figure 5).
Figure 5. Comparison between the genomes of Henry and Cjw1
The comparison was obtained using Phamerator [9]. Gp34 of Henry and gp33 of
Cjw1 have an E-value of 0.0 and are from the same phamily. Therefore, gp34 of
Henry may also be a putative holin protein based upon homology with the putative
holin protein (gp33) of Cjw1.
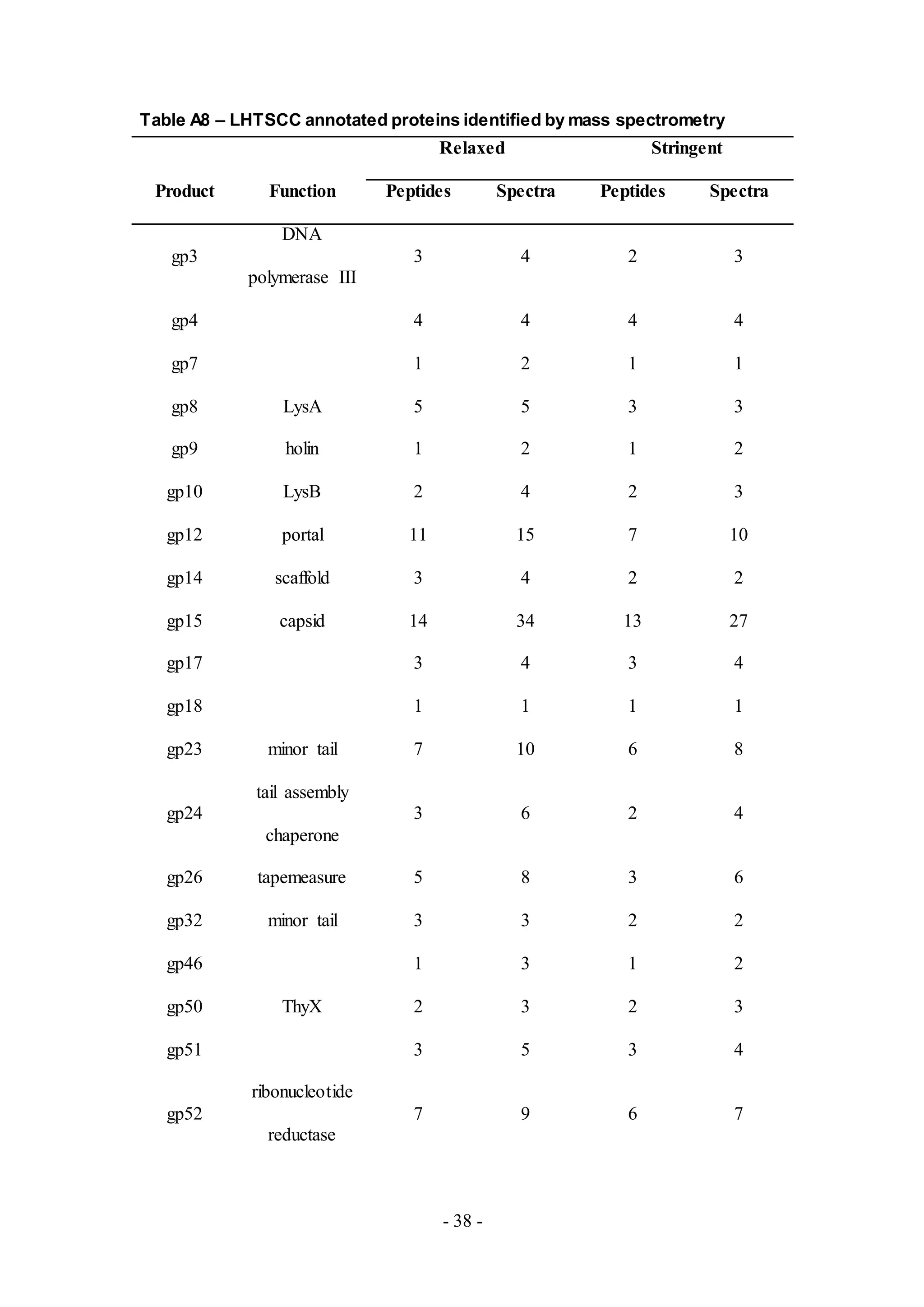
Twenty one proteins from LHTSCC (Table A8), a Cluster A4 phage, were detected,
including those with putative functions in DNA replication, virion structure and
assembly and lysis, similar to observations in other phage samples. Unique proteins
not detected in other phage samples were also observed, including a putative
recombination directionality factor (RDF) and ThyX (gp50) encoding flavin-
dependent thymidylate synthase. Interestingly, a peptide of the tapemeasure protein](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-9-2048.jpg)
![- 10 -
(gp26) covering an additional six amino acids upstream from the annotated sequence,
was detected as well (Figure 6).
Figure 6. A peptide of LHTSCC tapemeasure protein (gp26) covering six amino acids
upstream from the annotated start site.
The green bars are annotated putative genes, and the pink bar is the detected peptide
matching six amino acids upstream from the annotated tapemeasure protein (gp26).
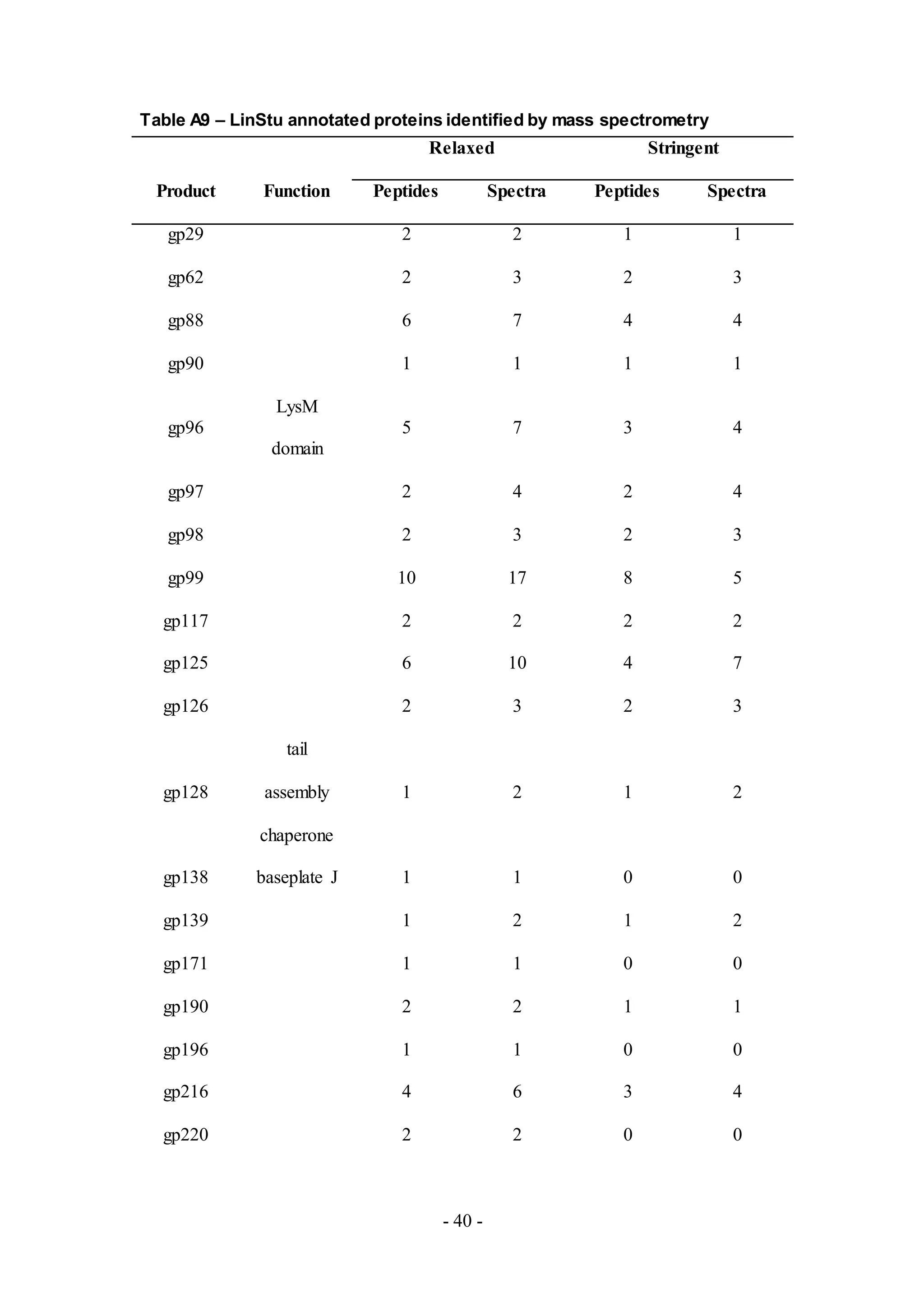
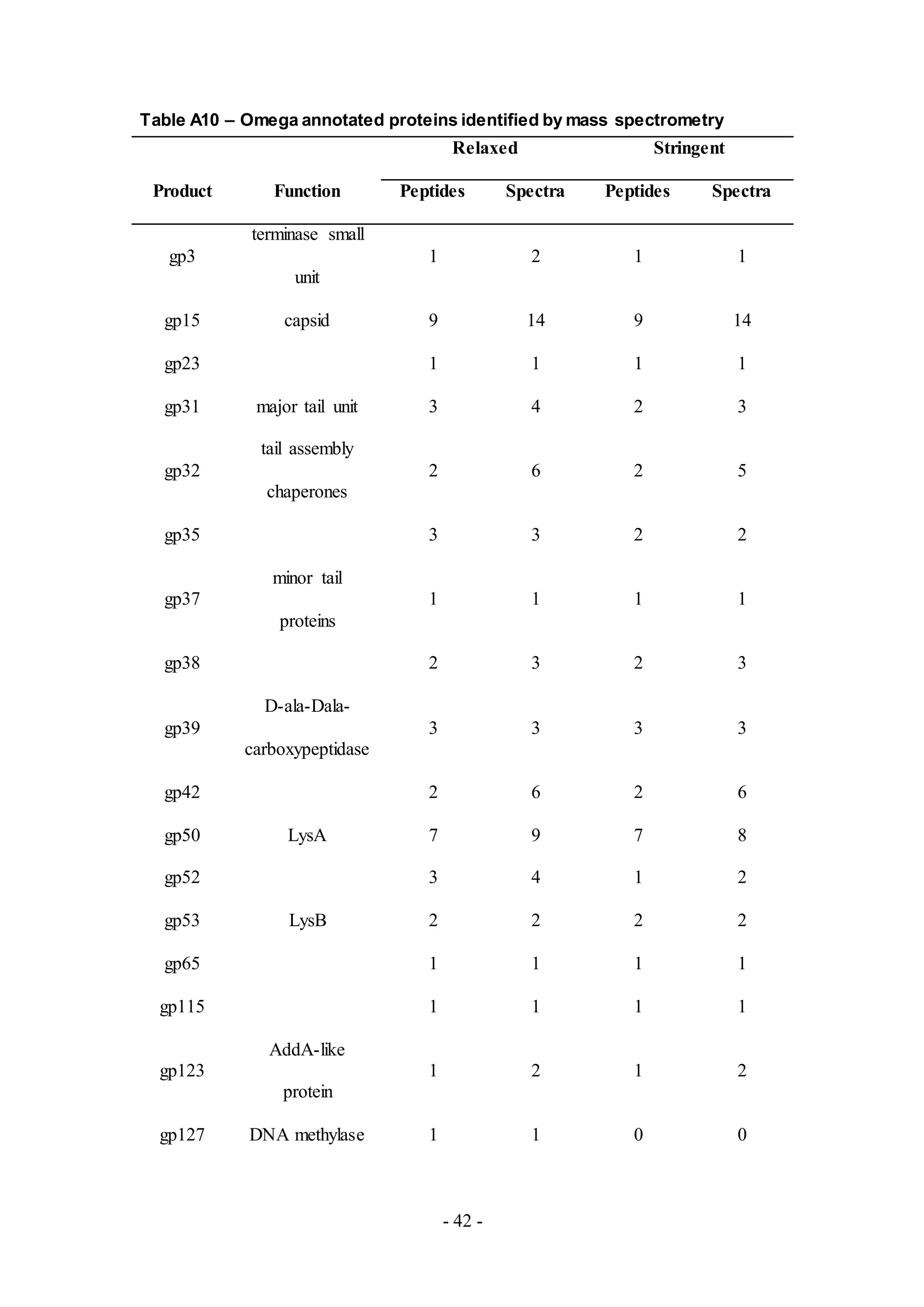
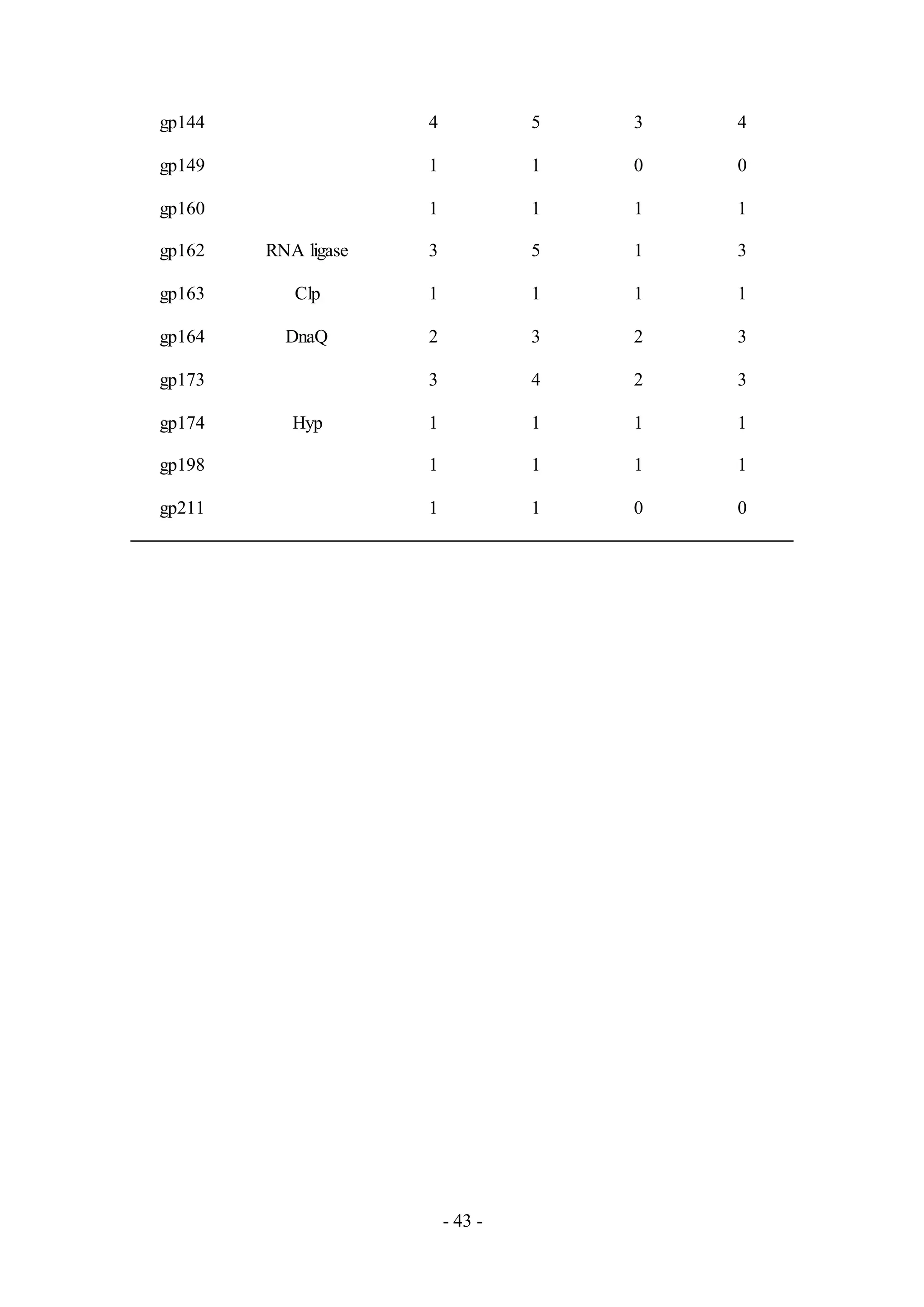
Similar to other phage samples, most of the peptides detected in the remaining phage
samples LinStu (Cluster C1, Table A9), Omega (Cluster J, Table A10), Papyrus
(Cluster R, Table A11), and Wee (Cluster F1, Table A12), were virion structure and
assembly proteins. Other proteins involved in DNA replication and lysis were also
detected in the phage Omega. The functions for the remaining proteins detected were
not characterized, as in the phage Papyrus where the putative functions of thirteen of
the eighteen proteins detected in the phage sample were unknown.
Identification of phage peptides using Mascot
Mascot is another common software that is used to analyze raw MS peptide data [6].
Thus, as an alternative to the data analysis process with X! Tandem, both Mascot and
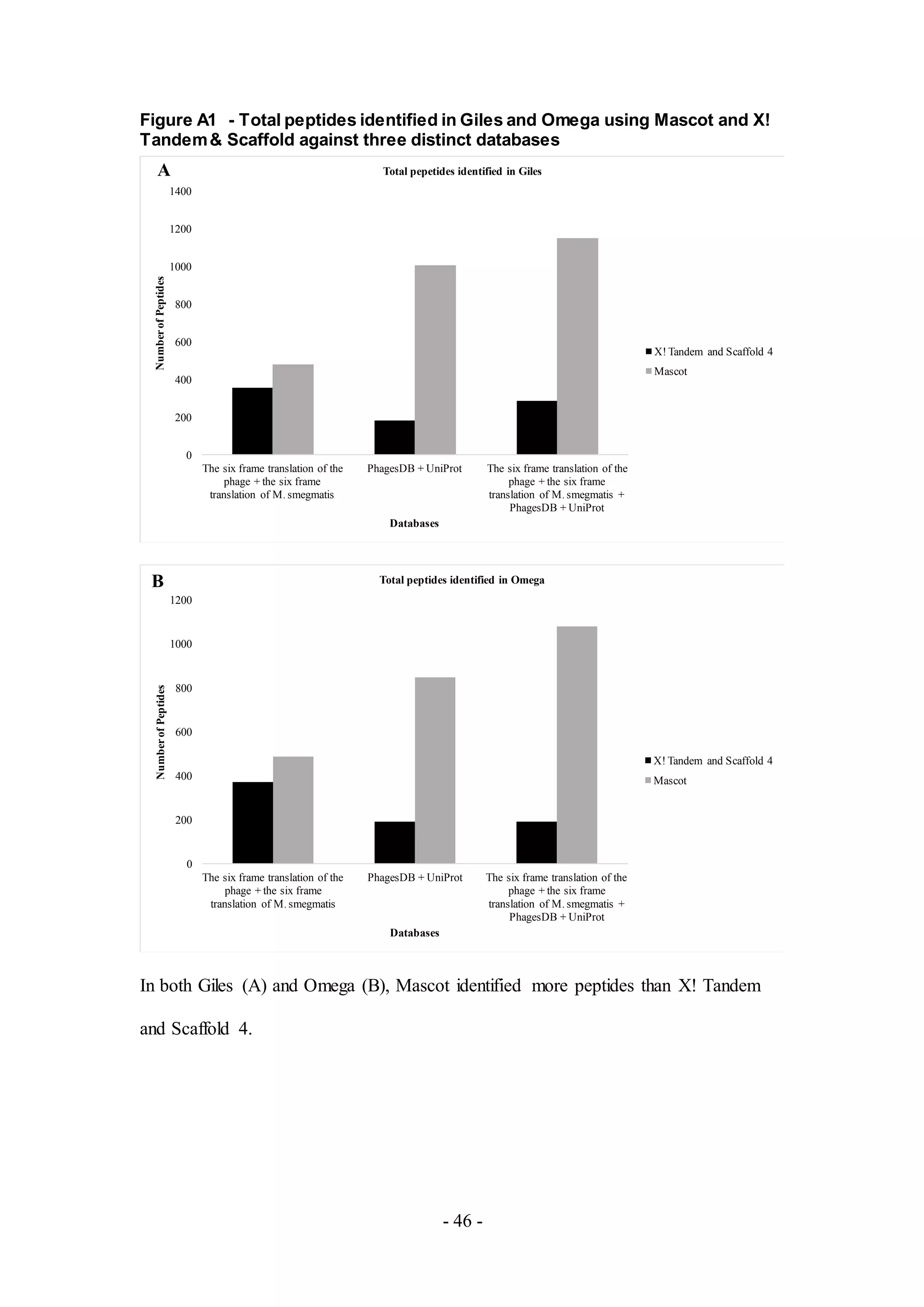
X! Tandem were used to search the mass spectra raw data of Giles and Omega against
three distinct databases: the six-frame translation of the sample phage and the six
frame translation of M. smegmatis, a public phage protein database PhagesDB and a
comprehensive protein database UniProt, and the combination of the six-frame
translation of the sample phage, the six frame translation of M. smegmatis, PhagesDB,
and UniProt. Based upon the results (Figure A1), Mascot identified more peptides](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-10-2048.jpg)
![- 11 -
than X! Tandem and Scaffold 4 with the same database and the larger database
resulted in more identified peptides. Thus, Mascot was used for cluster identification.
Mascot was used to search the mass spectra raw data against the public phage protein
database, PhagesDB [6]. As shown in Table 2, the range of peptides detected was
from 24 to 414 with over one hundred peptides obtained for phages Babsiella, Chah,
Giles, Henry, LHTSCC, LinStu, and Omega. Less than one hundred peptides were
detected in the remaining samples of phage. Searching against the PhagesDB database
resulted in the identification of peptides from multiple phages, and in some samples,
Chah, Henry, Lintsu and Wee, only a small portion or even none of the peptides
belonged to the sample phage (Figure 3). In contrast, over half of the total peptides
identified from Babsiella and Papyrus belonged to the predicted putative proteins
from these phages.
Figure 3. Percentage of the peptides from the sample phages in the total peptides
identified. Only a small portion of the identified peptides belonged to the predicted
putative proteins from the sample phages.
Identification of phage cluster
Clustering phages as described earlier and characterizing the putative proteins in
novel phages, currently relies on genomic DNA sequencing, which can be costly and
time-consuming. Previous studies have employed rapid protein extraction with mass
spectrometry to quickly identify microorganisms, such as bacteria and fungi, for
laboratory-based diagnostics [10, 11, 12]. Thus, it may be possible to categorize new
phages by analyzing the similarity between isolated phage peptides and annotated](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-11-2048.jpg)
![- 12 -
putative phage proteins in database PhagesDB [13], since phages are clustered based
upon nucleotide similarity [14] and putative phage proteins are predicted from the
genomic DNA.
Peptide identification using Mascot may result in a match to peptides of homologous
proteins in other phages since all of the archived putative proteins in PhagesDB are
searched. These homologous proteins may be from the same phamilies and include
proteins belonging to phages from various clusters [4]. Since the phages from the
same cluster have the most similar nucleic acid and predicted amino acid sequences, it
was predicted that most of the homologous proteins may belong to the phages from
the same cluster as the sample phage. Therefore, we decided to investigate if the
Mascot analysis results could be further utilized to categorize phages and predict their
cluster. The phage cluster that includes the largest number of detected peptides (the
most frequently detected cluster, the MFDC) in the output of Mascot analysis might
match the initial cluster assignment of the phage samples from genomic analysis.
The MFDCs of the phages span over 50% of all detected peptides (Table 3). Chah and
Henry had a MFDC that covered around 90% of the peptides detected, while LinStu
had a MFDC covering less than 70% of the peptides. Based upon the criteria, all the
phages listed in Table 3 had the MFDC that matched the initial cluster assignment
from nucleotide analysis.](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-12-2048.jpg)
![- 13 -
Discussion
Validation of phage genome annotations
Most of the peptides detected from the MS experimental analysis validated the
existing phage genome annotations of putative proteins. A more thorough validation
was limited, however, by the number of peptides detected and the overall coverage of
the translated genomic sequence. The matching peptides did not overlap with
annotated start sites for putative proteins with one notable exception: the identified
peptide from the tapemeasure protein (gp26) of phage LHTSCC matched six amino
acids upstream from the predicted annotated start site. This suggests that the actual
start site of gp26 may be different from the current annotated start site. Future
applications will explore strategies to optimize coverage and the number of phage
peptides isolated and subsequently detected and analyzed, by changing variables such
as the titer of phage, the incubation time before sample isolation, and the mixture of
protease used to digest the isolated proteins prior to MS.
Studying uncharacterized phage proteins produced by native expression
Previous studies used recombinant protein expression to characterize the function of
non-structural proteins [15, 16, 17]. The recombinant expressed proteins, however,
may differ from the native phage protein and this could impact function. Native phage
proteins may undergo unique posttranslational modifications including the addition
or removal of specific amino acid residues [18, 19, 20]. Since phage proteins perform
specialized functions in host bacterial cells, it is likely that native phage proteins have
certain properties that facilitate functional interactions with host bacterial proteins. In
fact, recent research [21] revealed that the protein encoded by lysA of phage D29, is
only active in M. smegmatis and becomes inactive in Escherichia coli (E. coli). The](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-13-2048.jpg)
![- 14 -
method designed in this research provides an effective way to study uncharacterized
phage proteins through native expression in the bacterial host cells.
Phage proteins, phage life cycles and bacterial physiology
The mechanisms that control phage protein expression are not well understood. Since
the phages do not have native metabolism, phage protein expression relies on the
machinery of the host bacteria. As the physiology of the host changes, the pattern of
protein expression for the phage is also likely to change in response.
Previous research showed that wildtype E. coli cells, infected with a single phage, or
with multiplicity of infection (MOI) of one, will predominantly be lysed; if MOI is
higher than two, the lysogenic cycle is preferred over the lytic cycle [22, 23]. It was
reported that the frequency of the lysogenic cycle would be increased in an
environment that was not favorable for bacterial proliferation [24].
The studies above suggest that both M. smegmatis cell culture growth phases and
MOI could affect the phage life cycle. In order to obtain a high frequency of the
phage lysogenic cycle, St-Pierre and Endy grew E. coli cell culture for phage
infection to stationary phase [25]. Pope et al investigated protein expression of the
mycobacteriophage Patience by infecting exponentially growing M. smegmatis, and
found that the peptide amount of some proteins change within the first 2.5 hours after
infection [7]. Future studies investigating changes in phage protein expression
isolated from infected bacterial cells over time, could provide a better understanding
of the putative phage proteins beyond previous methods focusing solely on purified
phage virion particles. A better understanding of the phage protein expression pattern
could help expand the applications of phage. Diverse applications could include: a](https://image.slidesharecdn.com/e03a8476-eb7a-4284-bc22-1dfb86e98a9e-160830201946/75/jbe_validation_MS_draft_6_12_16_clase-14-2048.jpg)