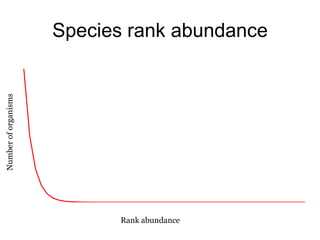
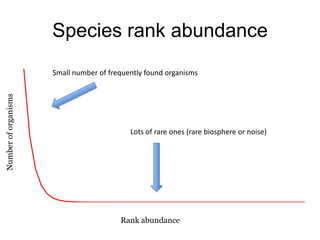
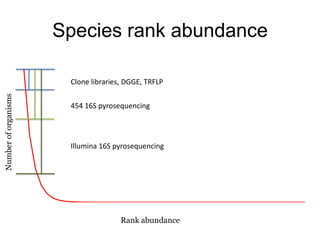
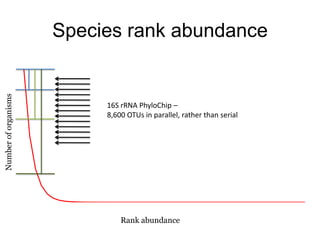
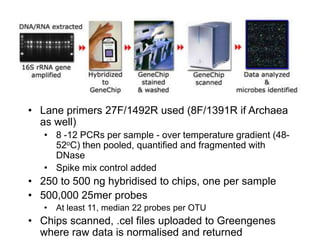
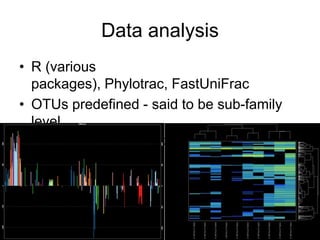
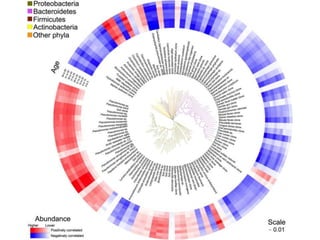
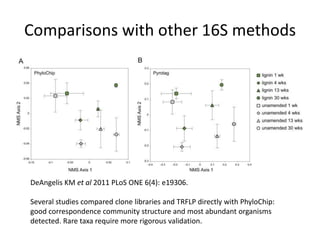
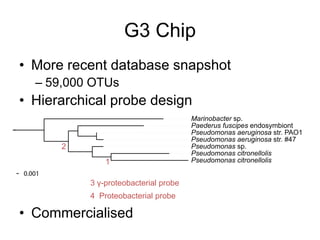
The document discusses the use of phylochip technology to characterize microbial communities in relation to human health and disease, focusing on its application in cystic fibrosis research. Despite identifying a significant number of operational taxonomic units (OTUs), the study found that only a few were strongly associated with known pathogens at varying ages. The phylochip method is compared to other sequencing methods, highlighting its efficacy in detecting community structures while noting the challenges associated with rare taxa and practical sampling considerations.