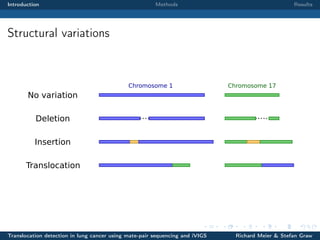
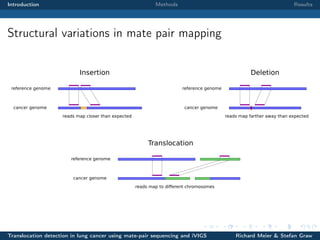
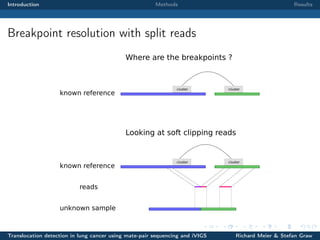
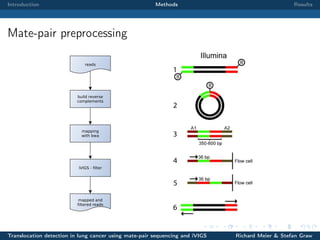
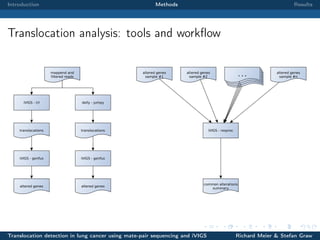
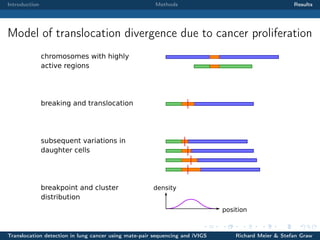
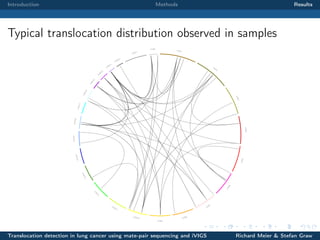
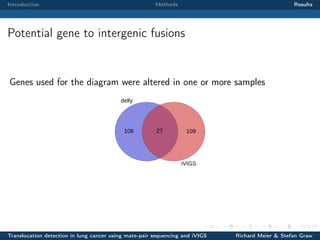
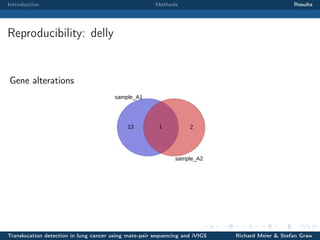
The document describes a study that used mate-pair sequencing and two structural variation detection tools (iVIGS and delly) to detect translocations in lung cancer samples. Mate-pair reads from 35 lung cancer patients were analyzed with iVIGS, while 32 samples were analyzed with delly. Both tools identified translocation breakpoints by clustering paired reads and using split reads. iVIGS estimated breakpoint positions with kernel density estimation, while delly reassembled split reads. Results varied between the tools. iVIGS showed better reproducibility and identified an average of 35 translocations per sample. Potential gene fusions involving known cancer genes were also identified. Future work will include validation of predicted gene fusions and examining