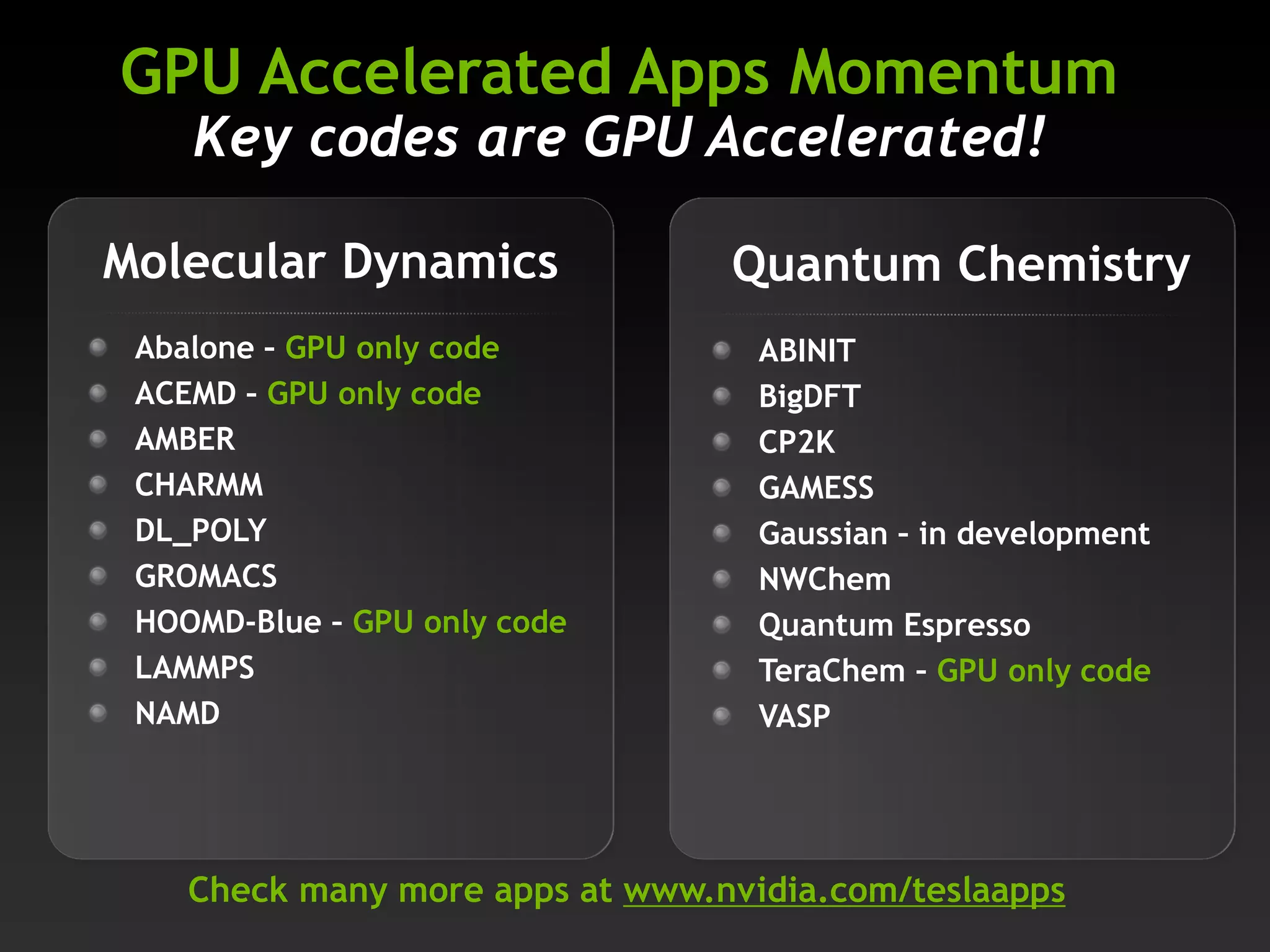
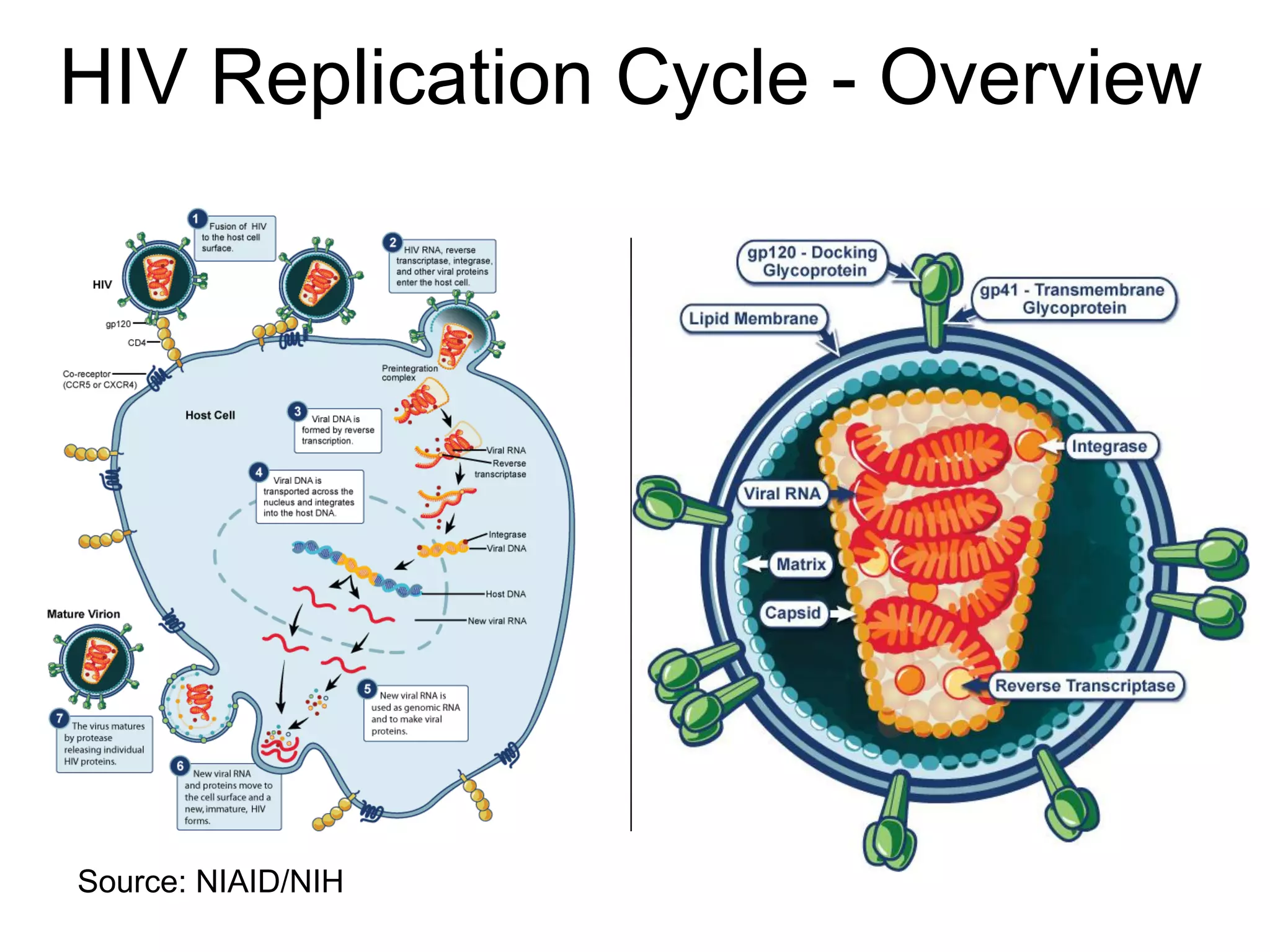
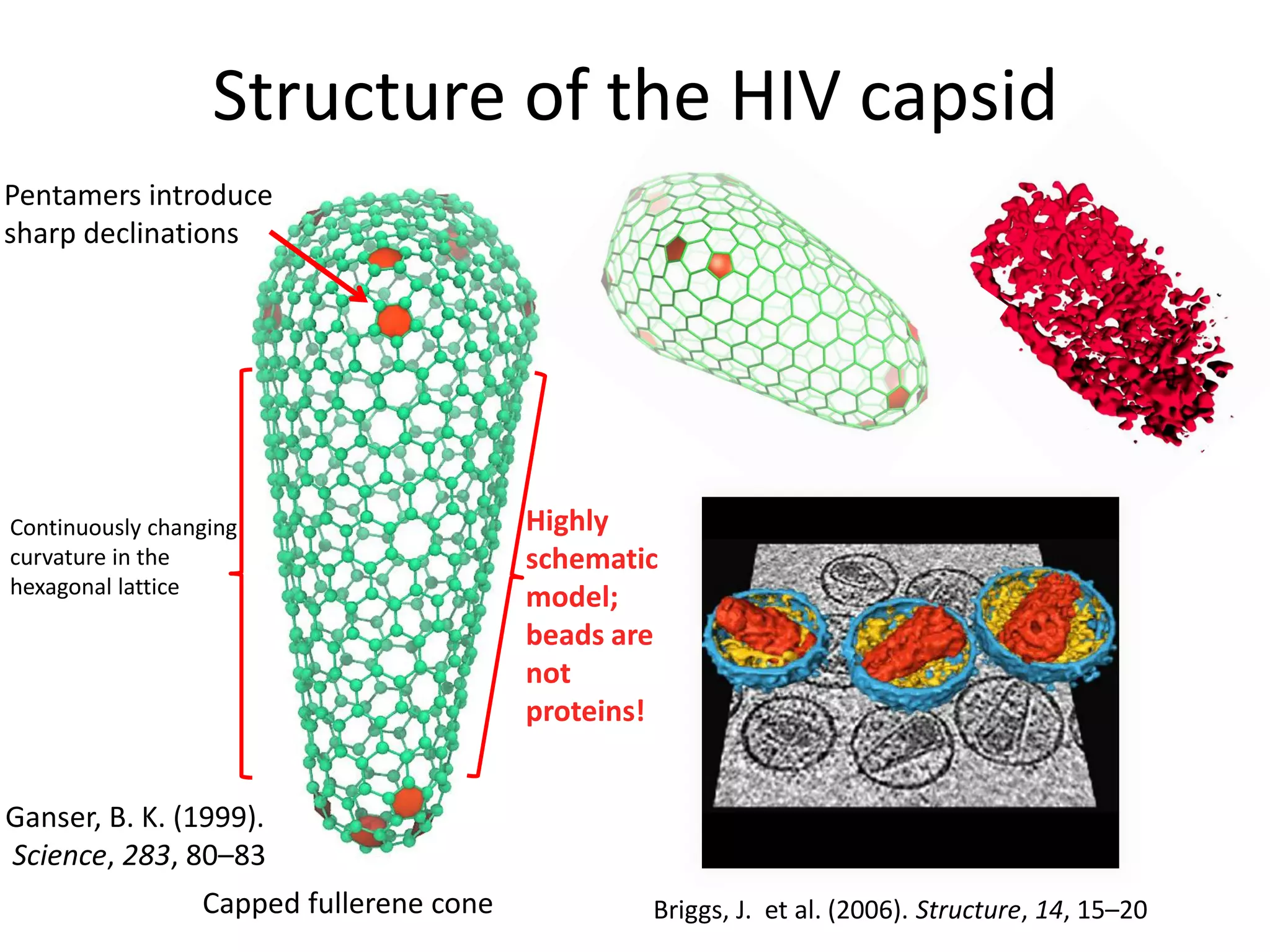
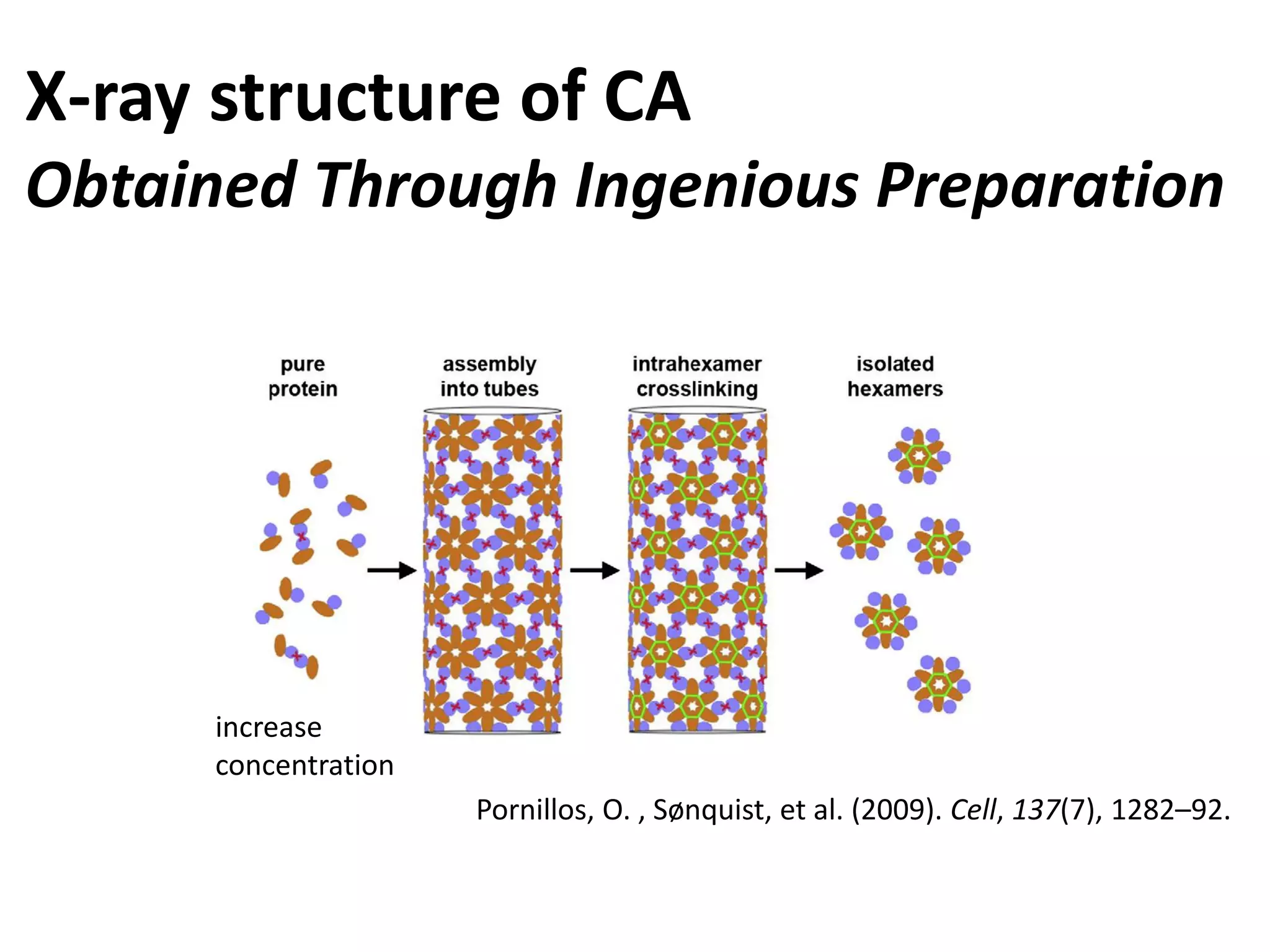
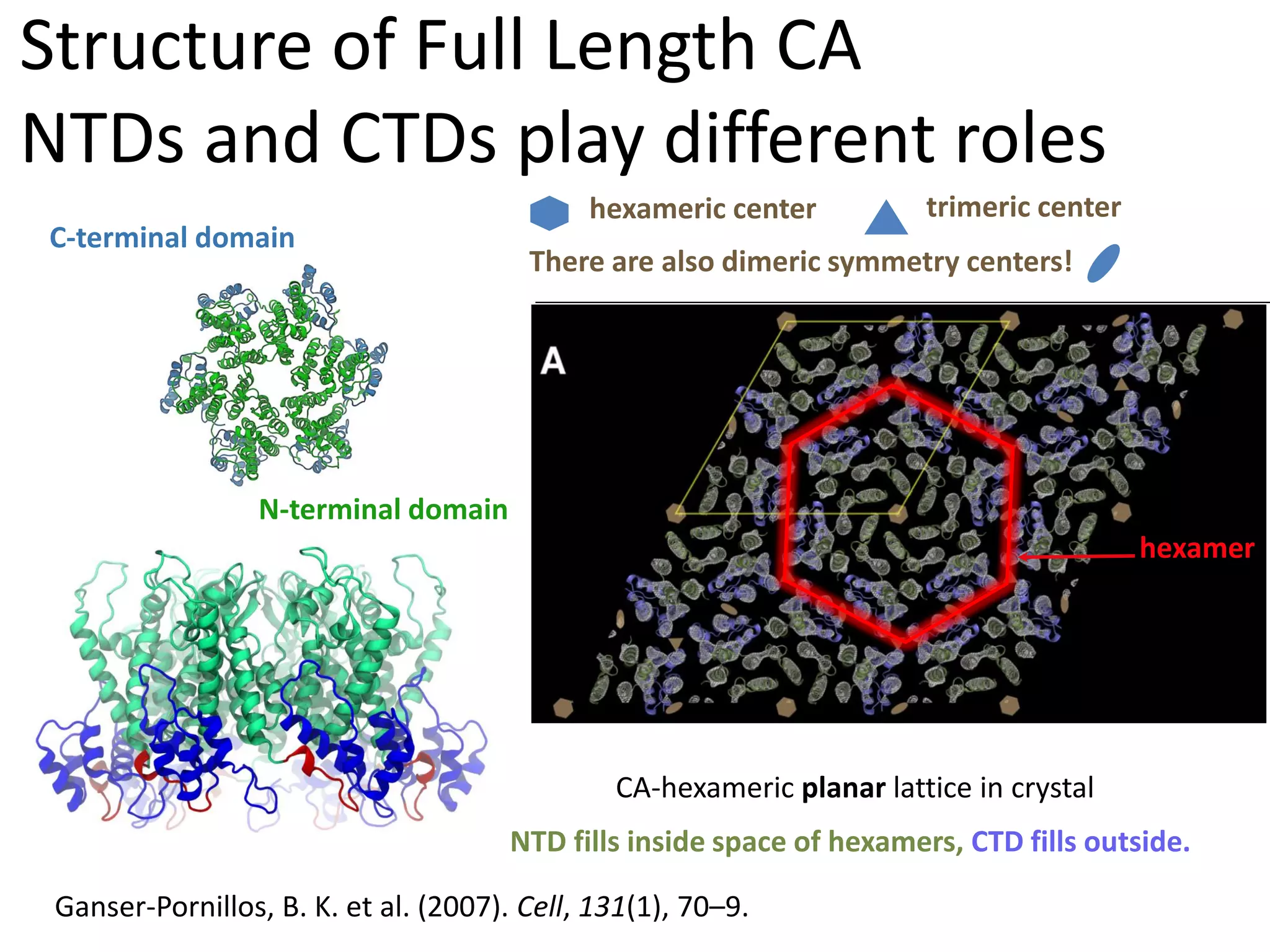
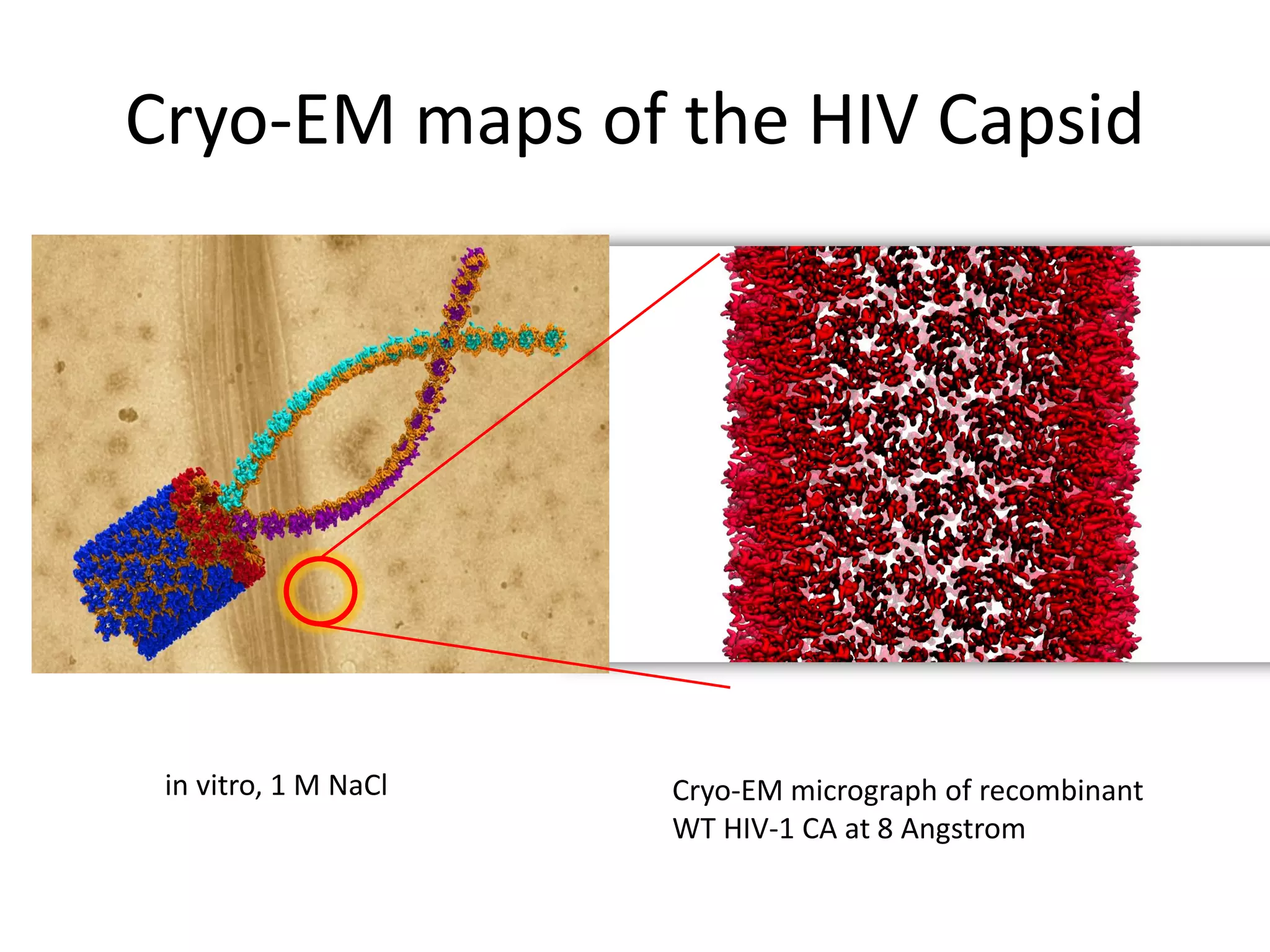
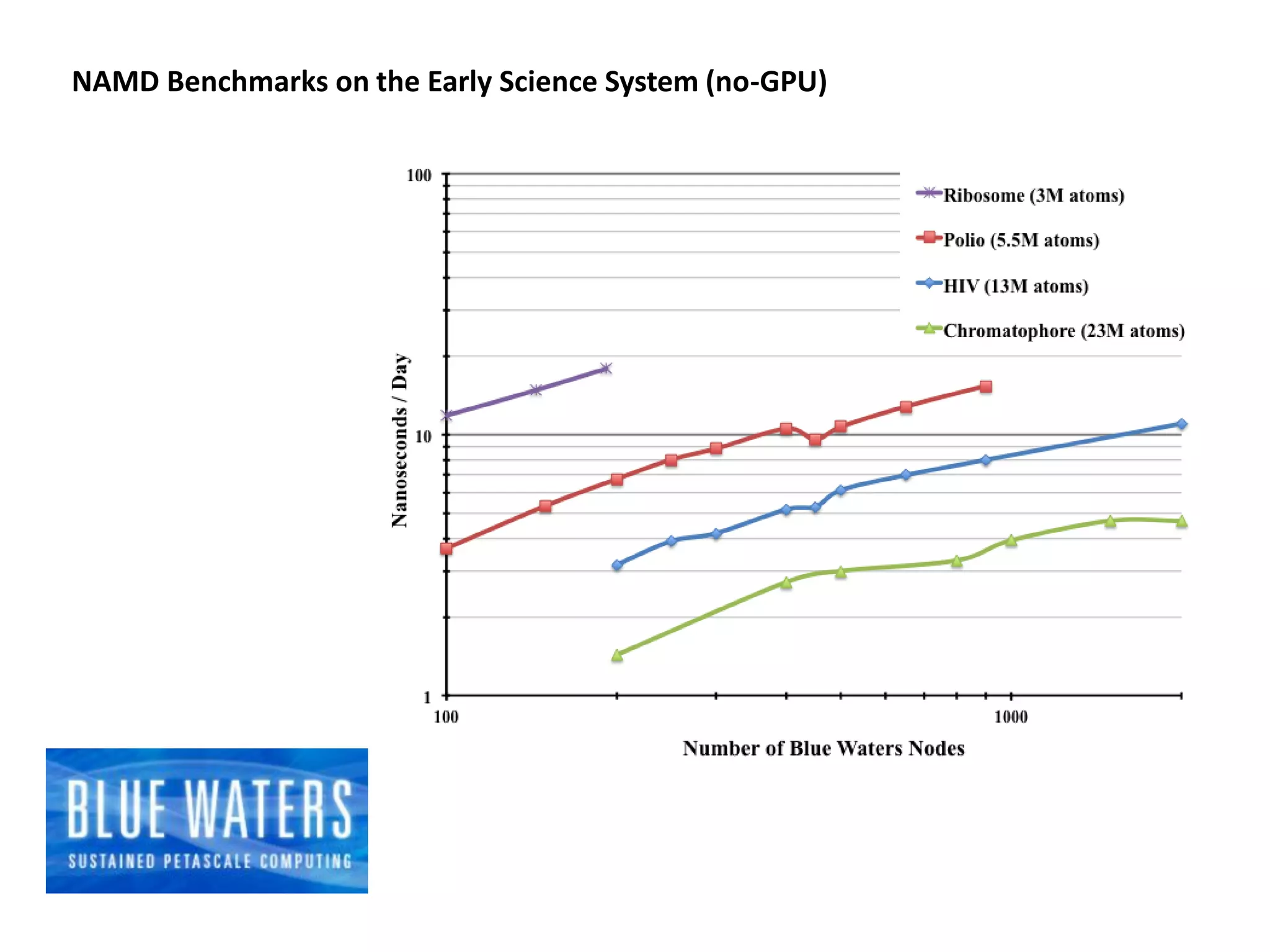
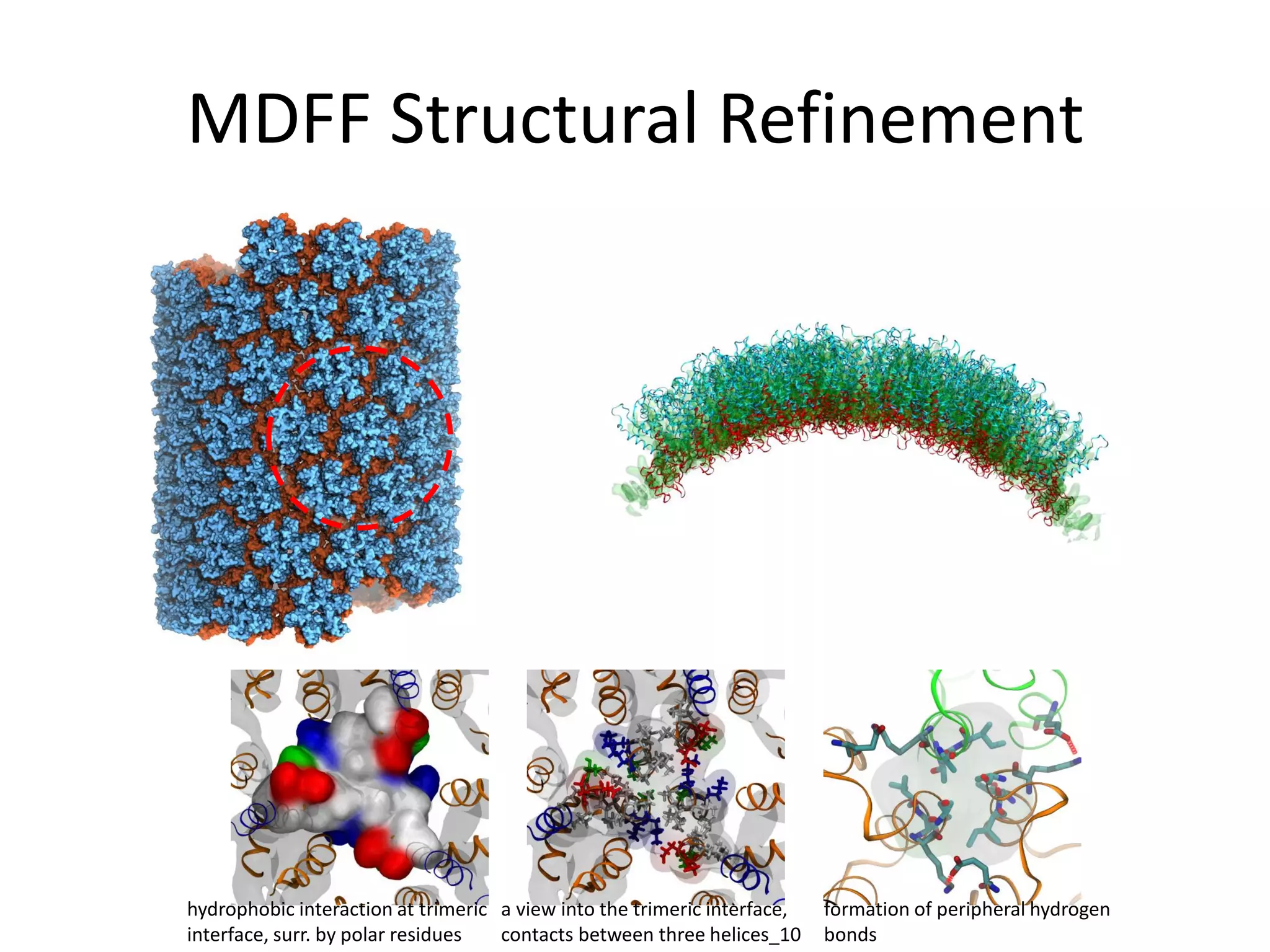
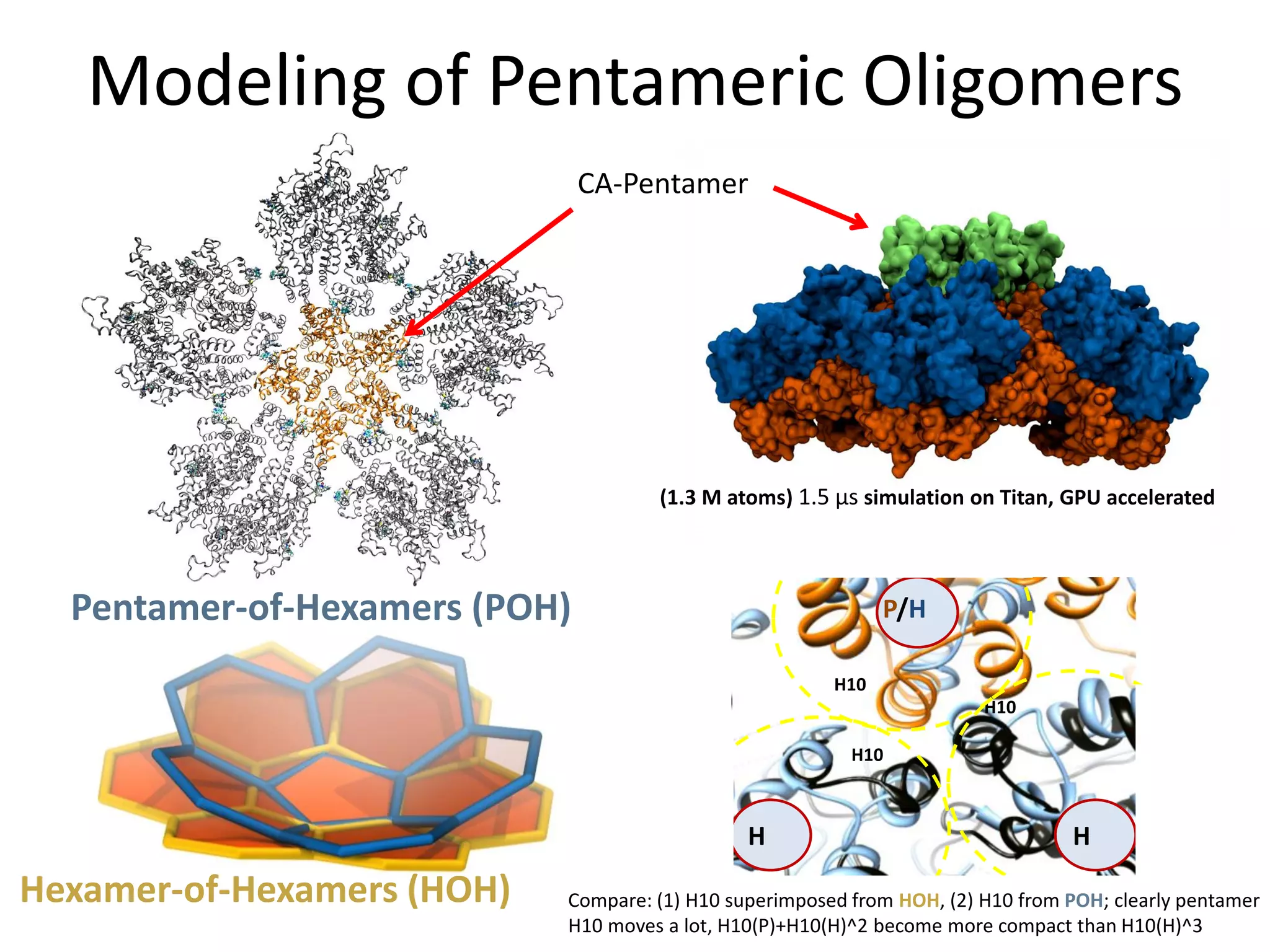
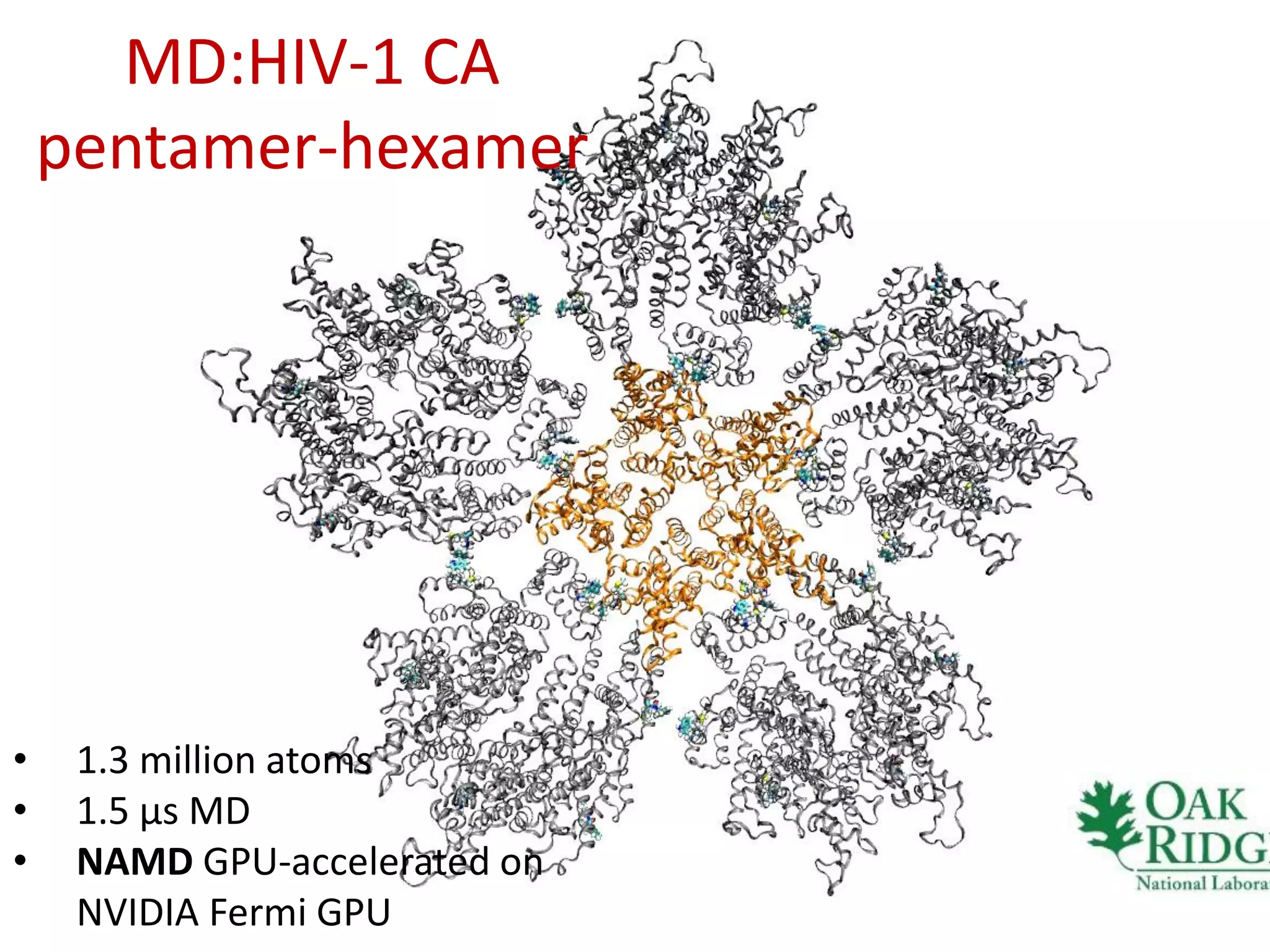
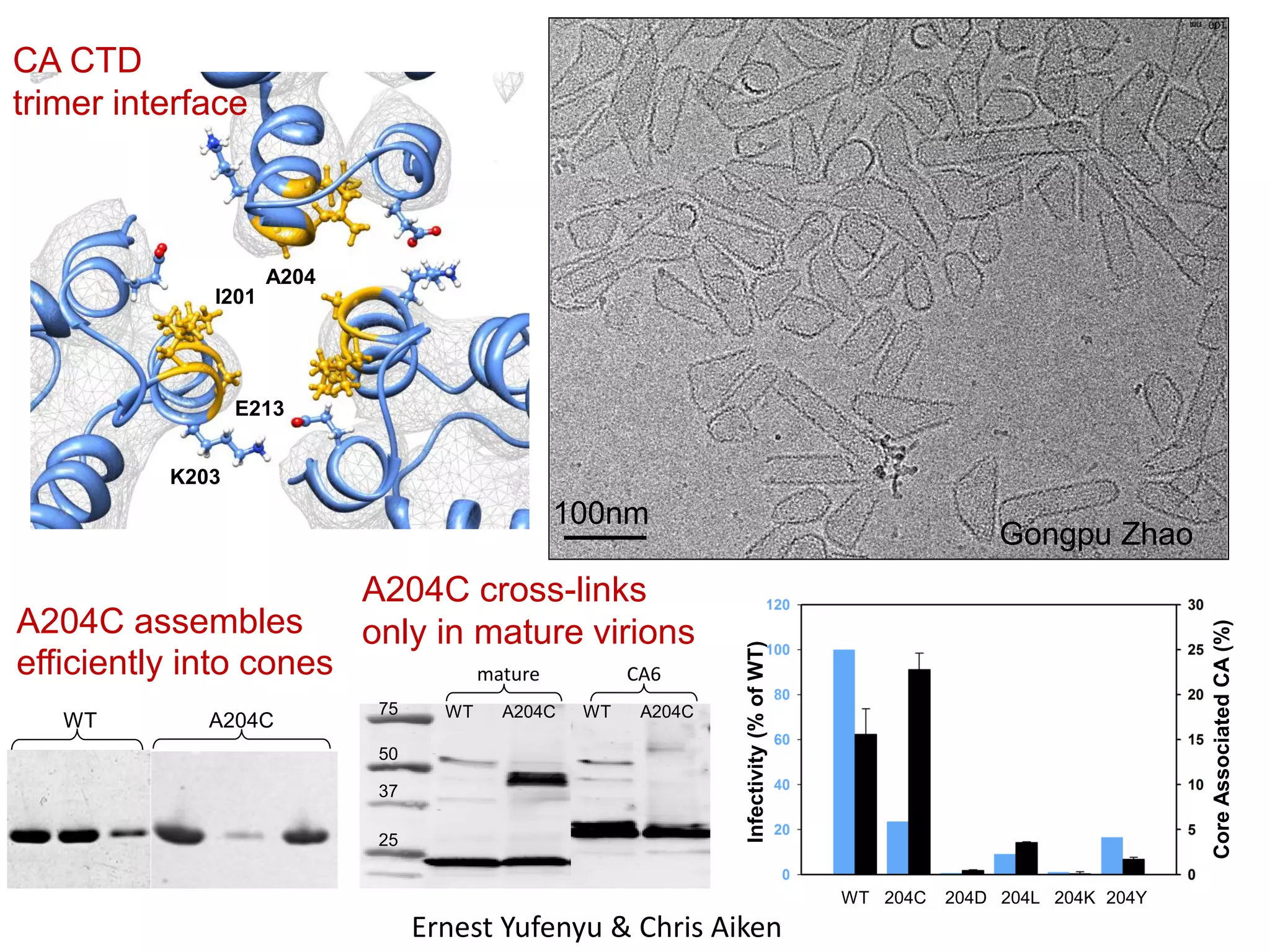
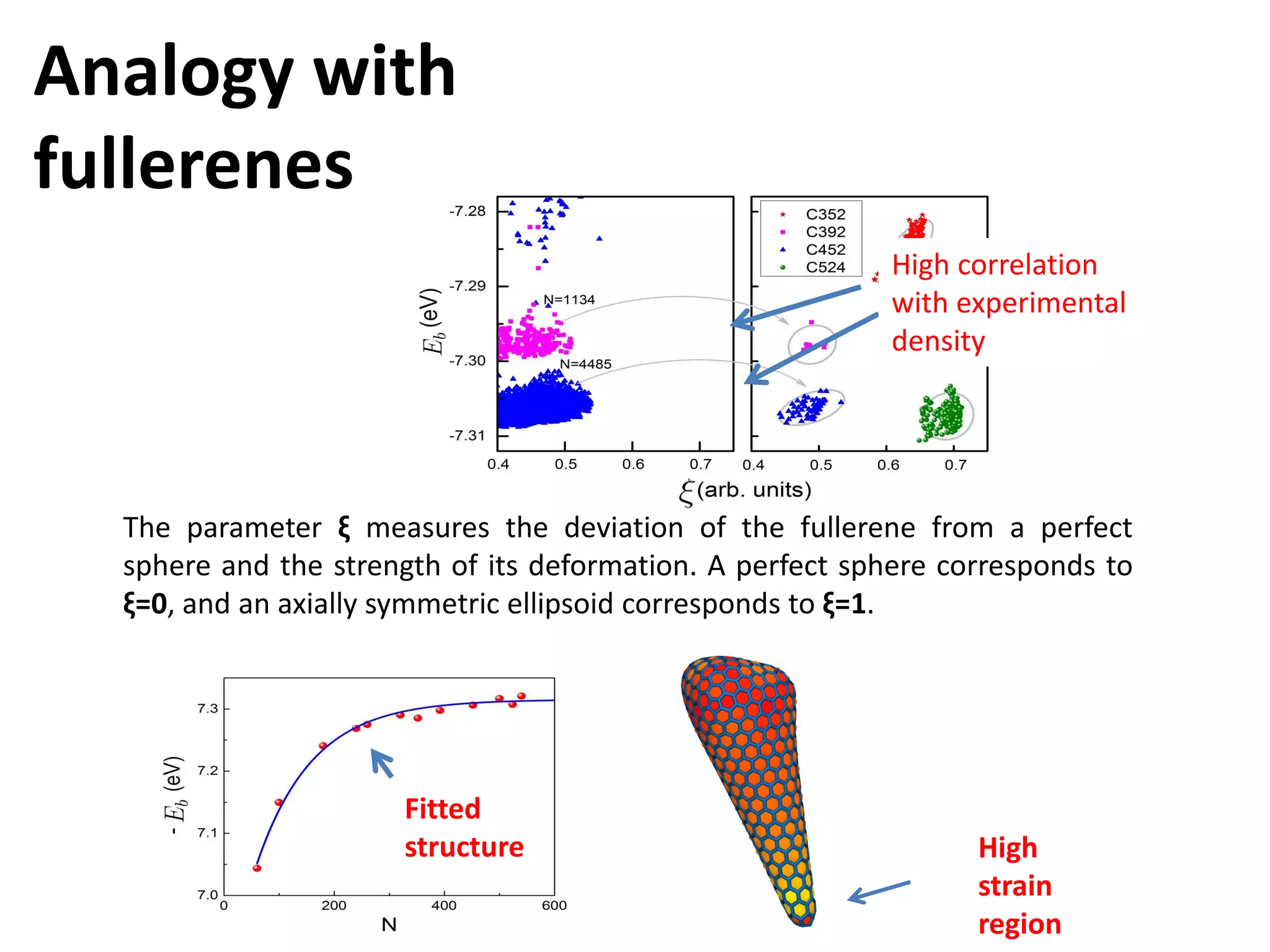
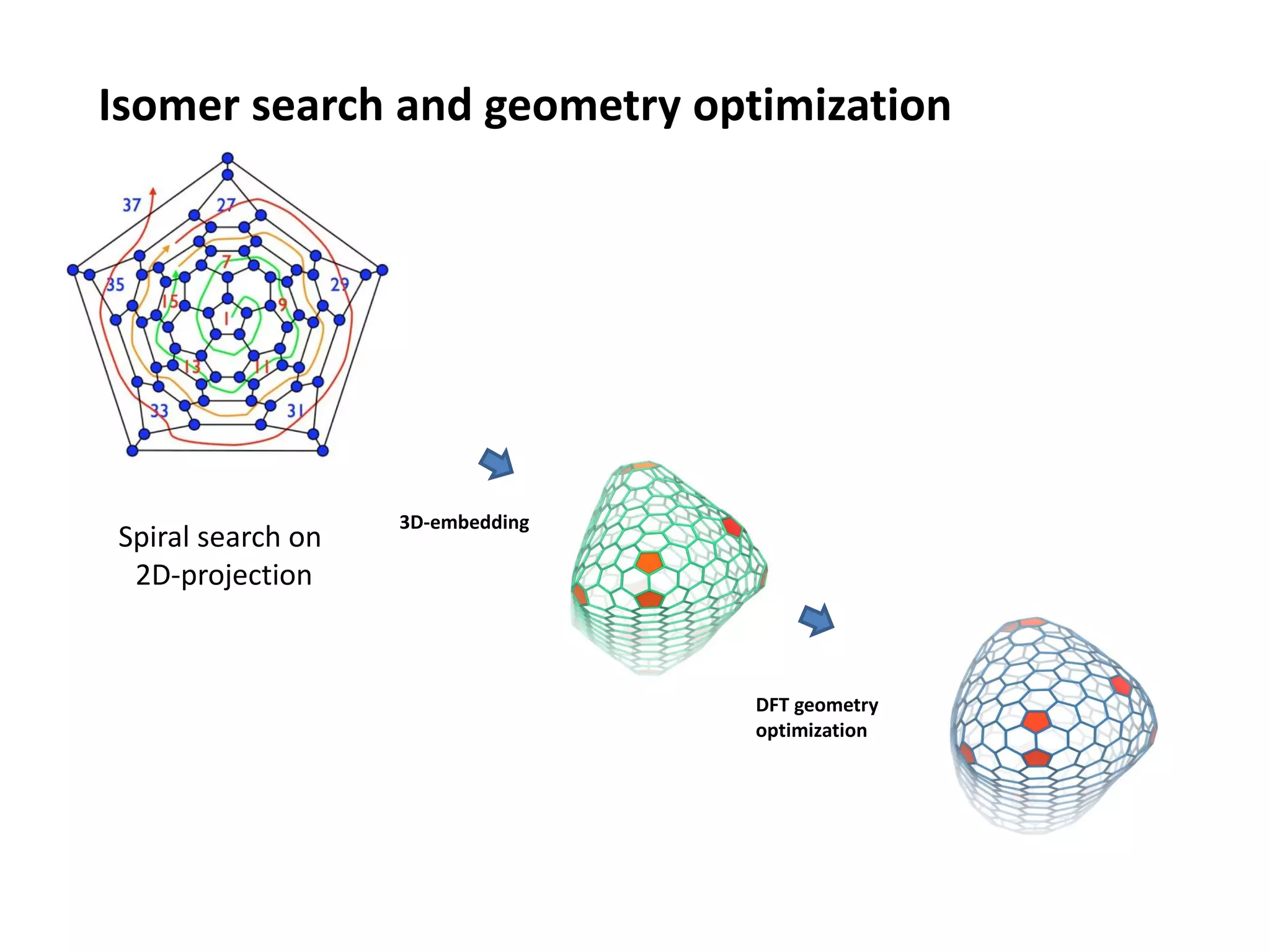
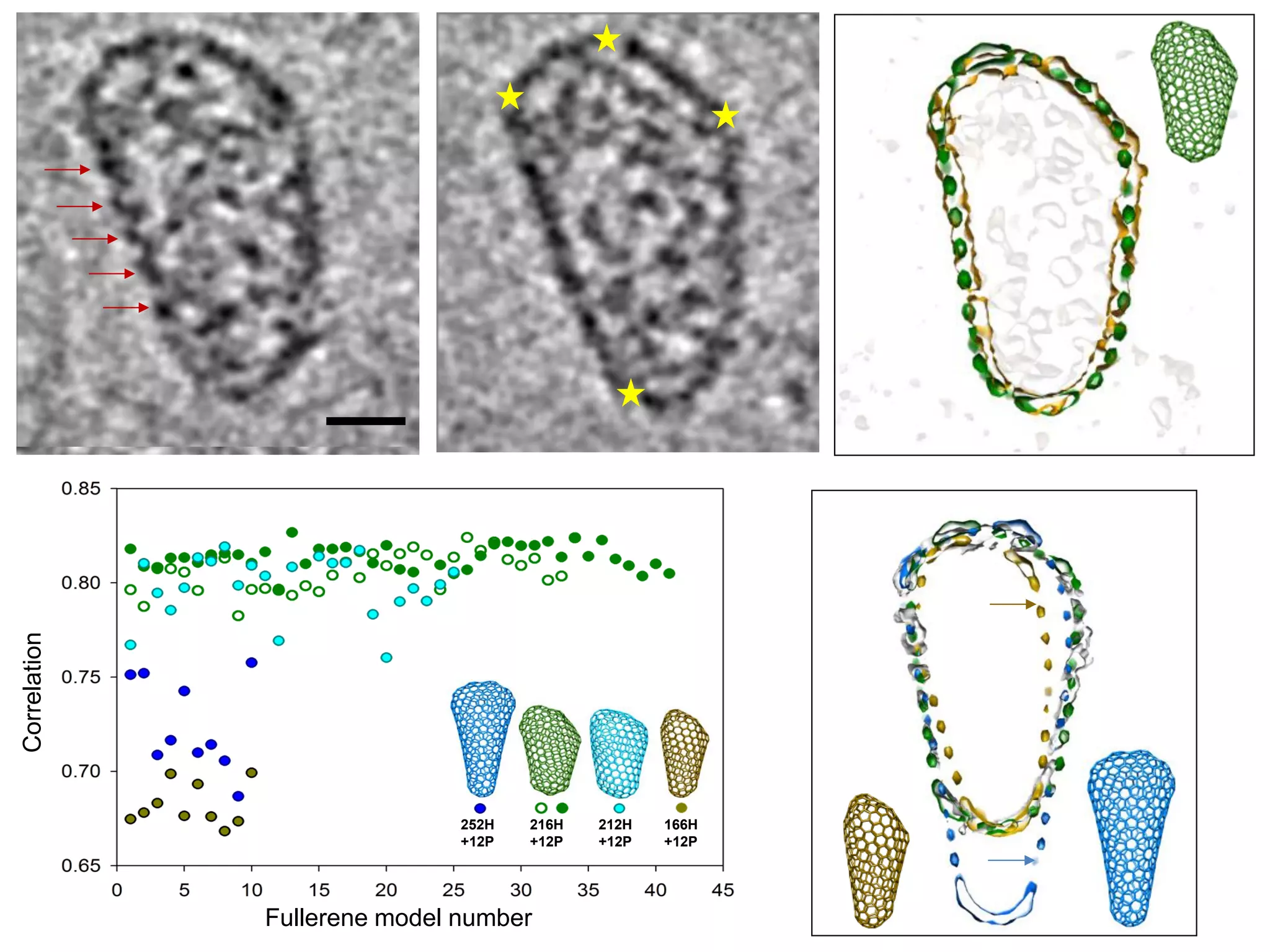
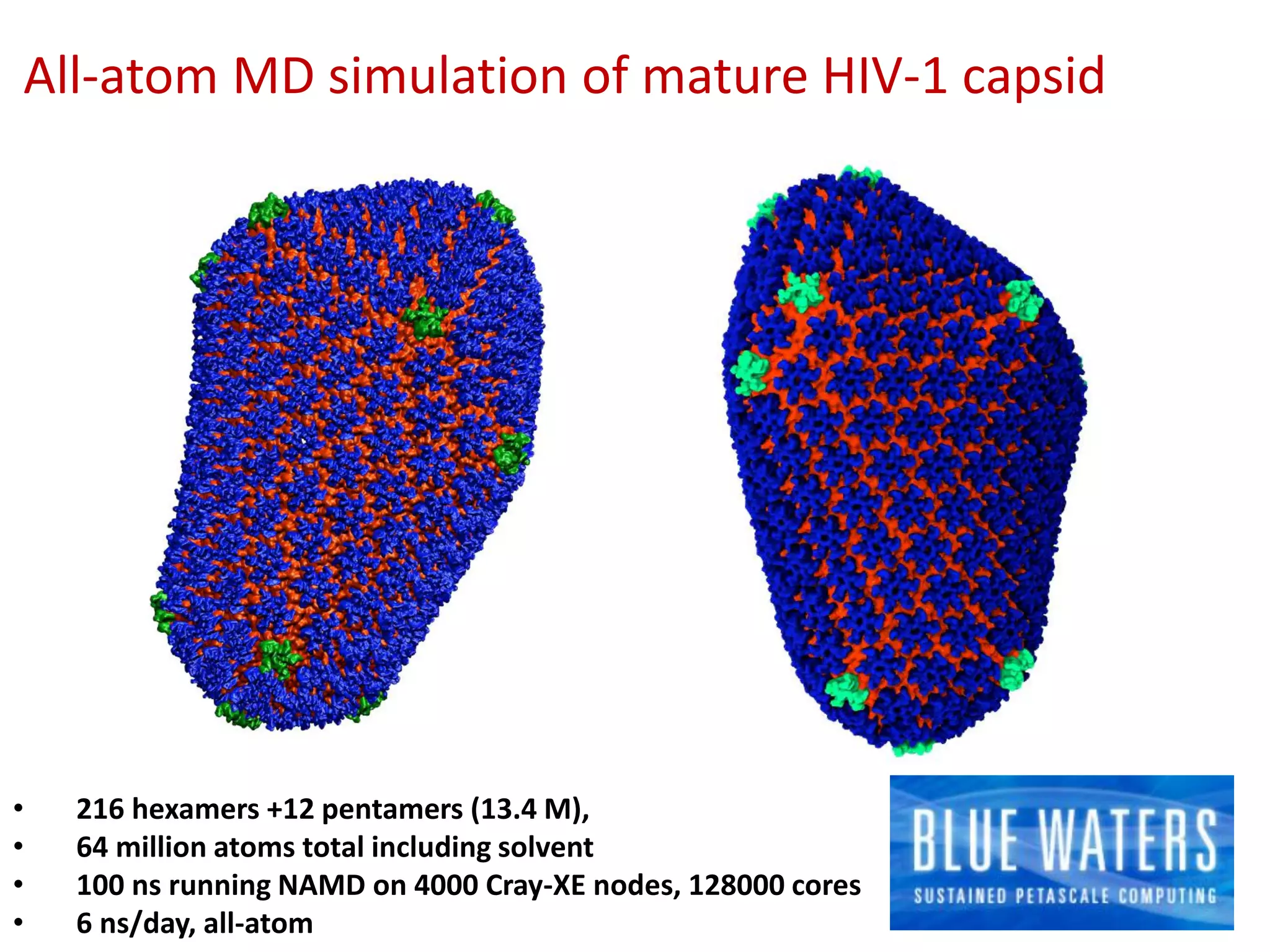
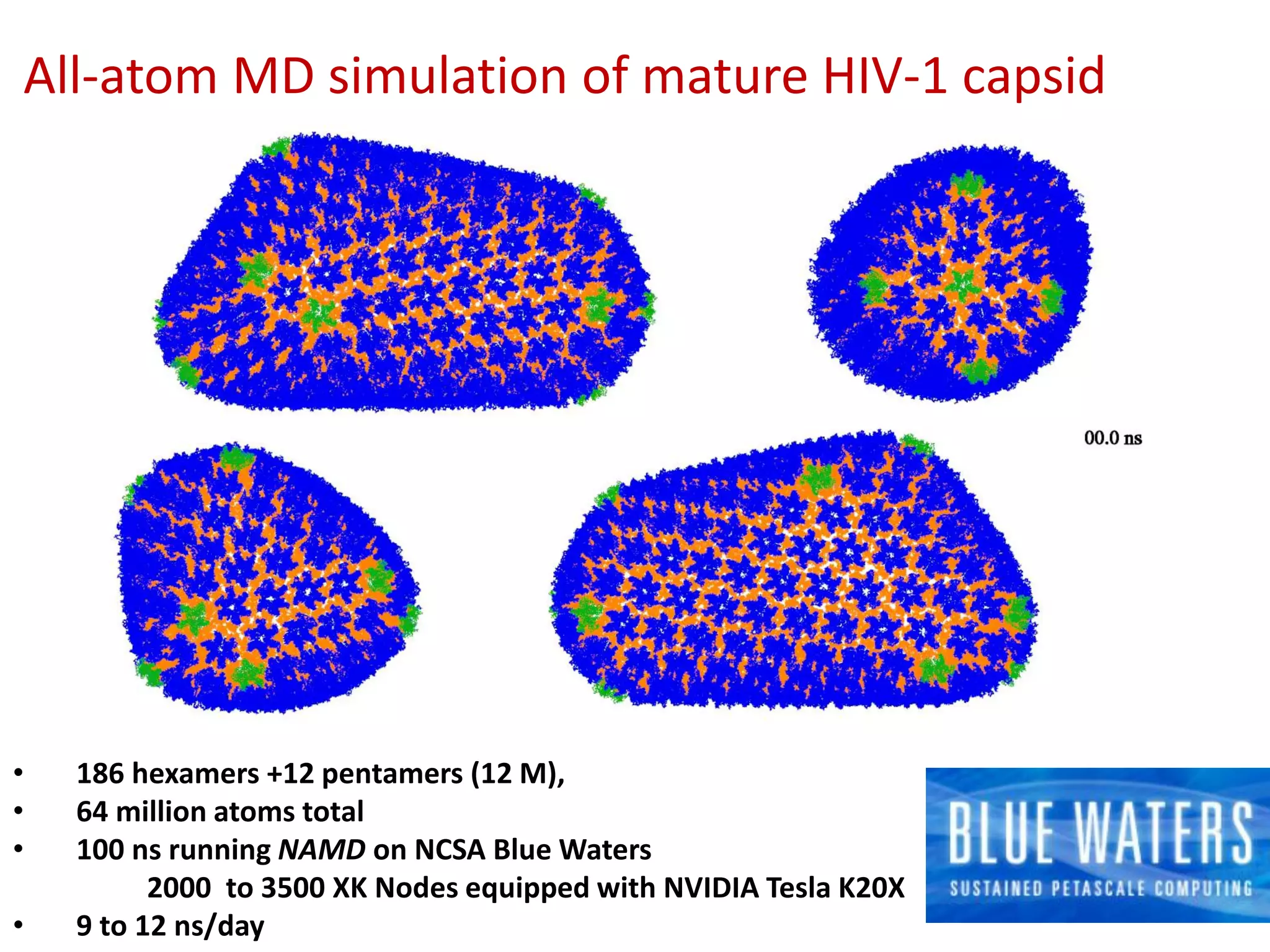
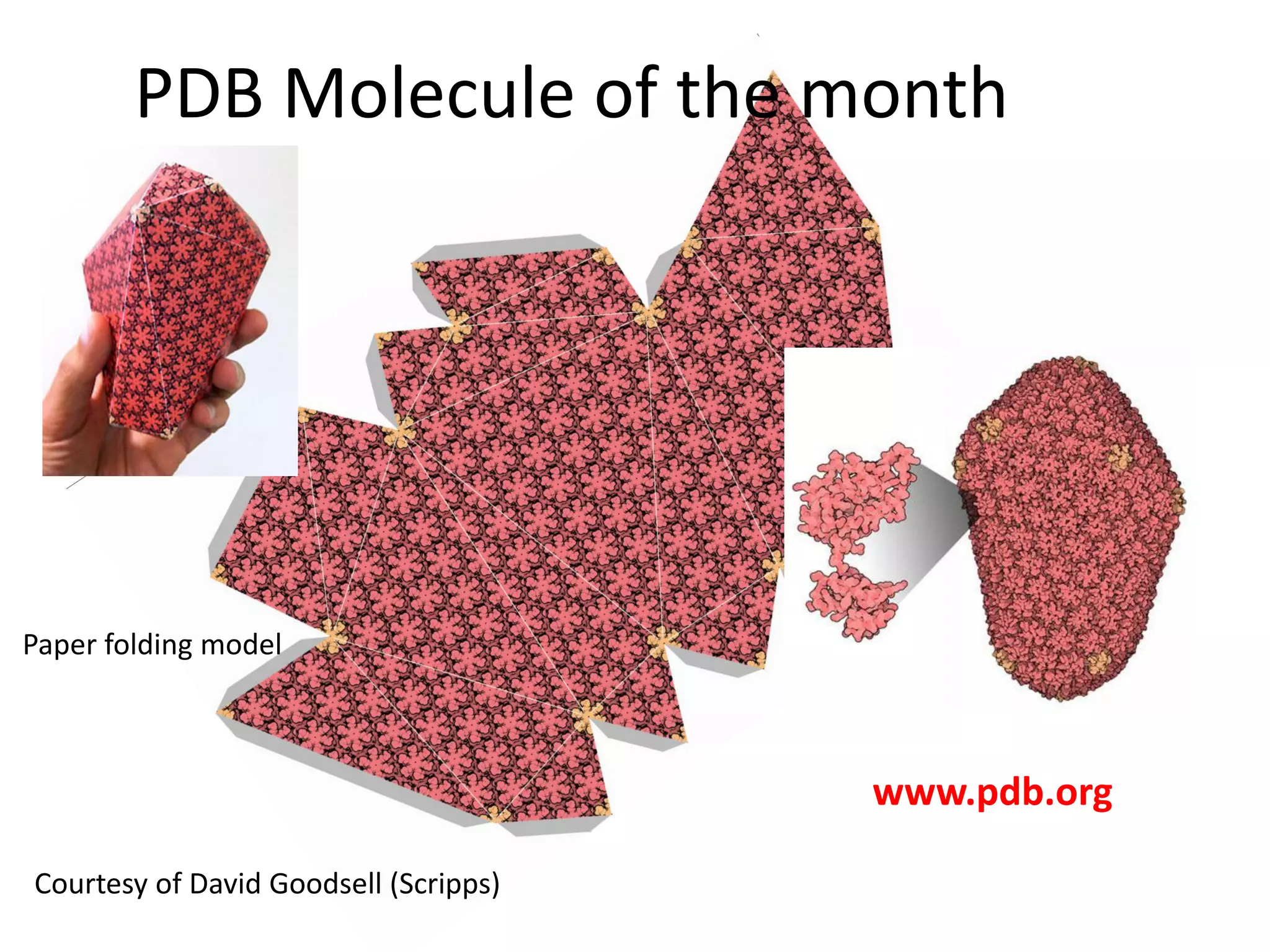
The document highlights various GPU-accelerated applications in molecular dynamics and quantum chemistry, emphasizing specific software codes optimized for NVIDIA GPUs. It discusses the structure and dynamics of the HIV-1 capsid, including detailed molecular simulations and findings relevant to its assembly and dynamics. Additionally, it includes information on upcoming webinars and opportunities for GPU test drives, promoting the use of NVIDIA technology in advanced research fields.