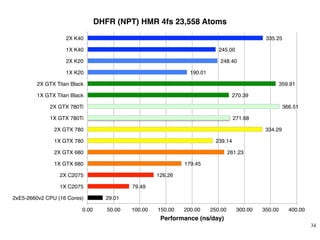
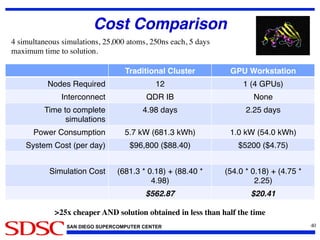
The document discusses the Amber 14 molecular dynamics (MD) simulation package and its new features, including GPU acceleration, improved performance capabilities, and expanded analysis tools. It highlights the collaborative development by various academic contributors and the availability of Amber and AmberTools as paid and free software respectively. Additionally, the document emphasizes the project's aim to make MD simulations more accessible and efficient for researchers, particularly through the use of commodity hardware.











































![[ ] [ ]
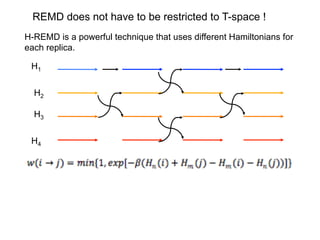
min(1,exp{( )( ( ) ( )})i j
m n E q E qρ β β= − −
vnew = vold
Tnew
Told
P X( )w X → X'
#$
%
' = P X '( )w X ' → X( )
P X( )= e
−βEX
Impose desired weighting (limiting
distribution) on exchange calculation
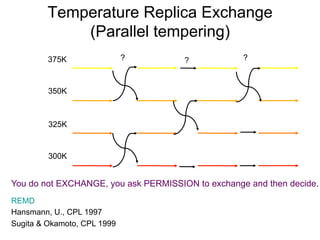
375K
350K
325K
300K
4 Impose reversibility/detailed balance
Rewrite using only potential energies, by rescaling kinetic energy for both
replicas.
Exchange?](https://image.slidesharecdn.com/20140511nvidiawebinaramber14-140515174950-phpapp01/85/AMBER14-GPUs-49-320.jpg)