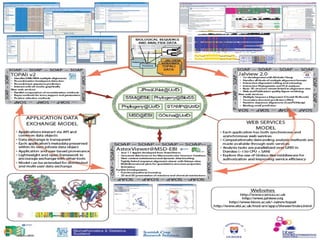
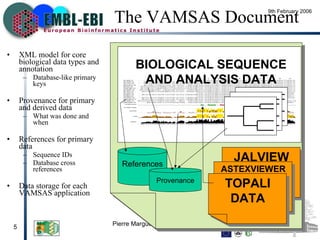
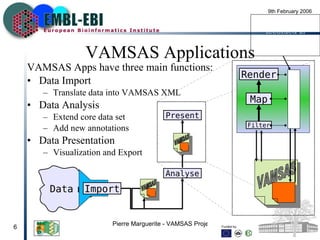
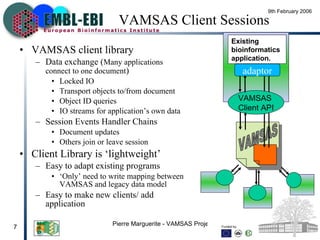
VAMSAS is an open framework that facilitates interoperation between advanced bioinformatics tools by providing a common data model. It brings together three programs - Jalview, TOPAli, and AstexViewer. The VAMSAS document XML format defines a model for storing core biological data types and annotations. VAMSAS applications import data, perform analysis to add annotations, and allow visualization and export of results.


![Bringing three programs together… The VAMSAS Framework Visual Analysis of Molecular Sequences, Alignments and Structures Jalview Alignment Visualization Sequence Analysis ( University of Dundee Geoff Barton & David Martin Jim Procter, Andrew Waterhouse ) TOPAli DNA Recombination Phylogenetic Analysis ( Biomathematics and Statistics Scotland (BioSS) At Scottish Crop Research Institute (SCRI) Frank Wright & David Marshall Iain Milne ) [email_address] Molecular Graphics Conformation Analysis Reaction Diagrams (Tom Oldfield & Kim Henrick Pierre Marguerite ) http://www.vamsas.ac.uk VAMSAS](https://image.slidesharecdn.com/groupmeeting-vamsasproject-final-090905031259-phpapp02/85/Group-Meeting-Vamsas-Project-Final-3-320.jpg)
![My work Integrates the AstexViewer@MSD-EBI in VAMSAS workflow. Conversion/Proceeding of Data Export from AV-MSD (annotations for other vamsas application) Separated/specific application (VAMAV) [email_address] [email_address] VAMSAS Client API adaptor](https://image.slidesharecdn.com/groupmeeting-vamsasproject-final-090905031259-phpapp02/85/Group-Meeting-Vamsas-Project-Final-8-320.jpg)
![Process of conversion Extract Alignment sequences and annotations Mapping with PBD ID Sequence Grouping Generate required files (atribute, alignment, …) Visualise data in AstexViewer@MSD-EBI VAMSAS Document XML – zip Sequence Grouping Generate required files Visualisation in [email_address] Document Processing](https://image.slidesharecdn.com/groupmeeting-vamsasproject-final-090905031259-phpapp02/85/Group-Meeting-Vamsas-Project-Final-10-320.jpg)