Feature scaling is a technique used in machine learning to standardize the range of independent variables or features of data. There are several common feature scaling methods including standardization, min-max scaling, and mean normalization. Standardization transforms the data to have a mean of 0 and standard deviation of 1. Min-max scaling scales features between 0 and 1. Mean normalization scales the mean value to zero. The document then provides the formulas and R code examples for implementing each of these scaling methods.








![R Code for arcsine transformation
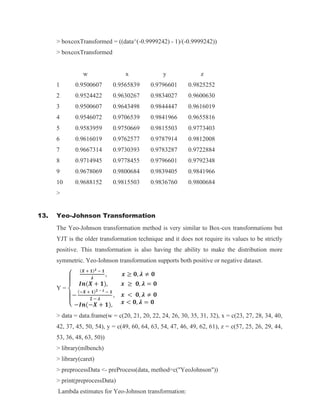
data = data.frame(w = c(20, 21, 20, 22, 24, 26, 30, 35, 31, 32), x = c(23, 27, 28, 34, 40, 42, 37,
45, 50, 54), y = c(49, 60, 64, 63, 54, 47, 46, 49, 62, 61), z = c(57, 25, 26, 29, 44, 53, 36, 48,
63, 50))
> arcsinemodified_w = data$w/max(data$w)
> arcsinemodified_x = data$x/max(data$x)
> arcsinemodified_y = data$y/max(data$y)
> arcsinemodified_z = data$z/max(data$z)
> arcsineTransformed_w = asin(sqrt(arcsinemodified_w))
> arcsineTransformed_x = asin(sqrt(arcsinemodified_x))
> arcsineTransformed_y = asin(sqrt(arcsinemodified_y))
> arcsineTransformed_z = asin(sqrt(arcsinemodified_z))
> arcsineTransformed = data.frame('w' = arcsineTransformed_w, 'x' = arcsineTransformed_x,
'y' = arcsineTransformed_y, 'z' = arcsineTransformed_z)
> arcsineTransformed
w x y z
1 0.8570719 0.7110504 1.065436 1.2570684
2 0.8860771 0.7853982 1.318116 0.6814770
3 0.8570719 0.8039209 1.570796 0.6976468
4 0.9154304 0.9165257 1.445468 0.7456738
5 0.9756718 1.0365703 1.164419 0.9894260
6 1.0389882 1.0799136 1.029336 1.1610142
7 1.1831996 0.9751020 1.011806 0.8570719
8 1.5707963 1.1502620 1.065436 1.0610566
9 1.2259397 1.2951535 1.393086 1.5707963
10 1.2736738 1.5707963 1.352562 1.0992586
10. Square Root Transformation
Square root transformation can be used as
[i] for data that follow a Poisson distribution or small whole numbers
[ii] usually works for data with non-constant variance
[iii] may also be appropriate for percentage data where the range is between 0 and 30% or](https://image.slidesharecdn.com/featurescalingwithr-221129025401-86be9876/85/Feature-Scaling-with-R-pdf-9-320.jpg)