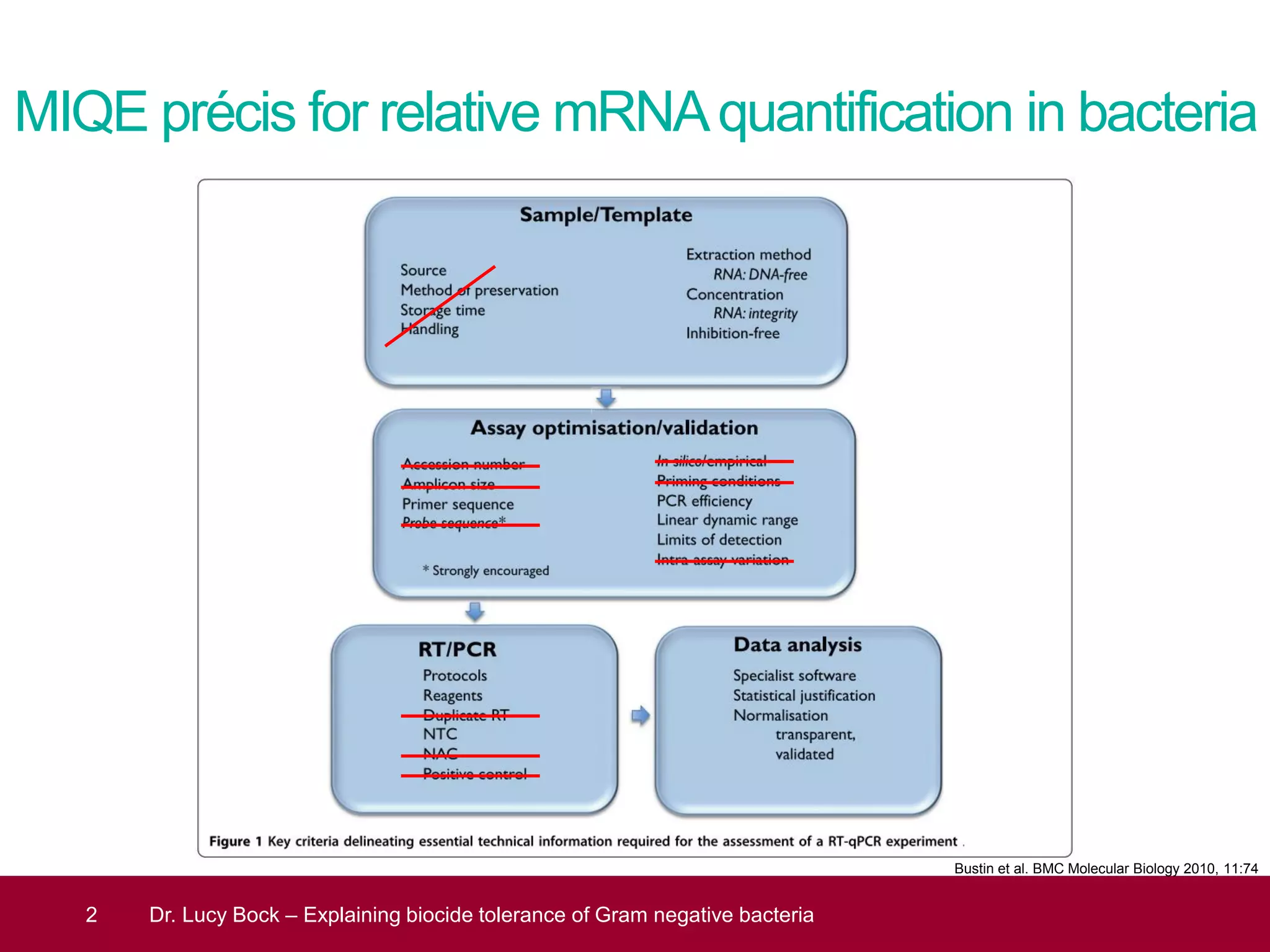
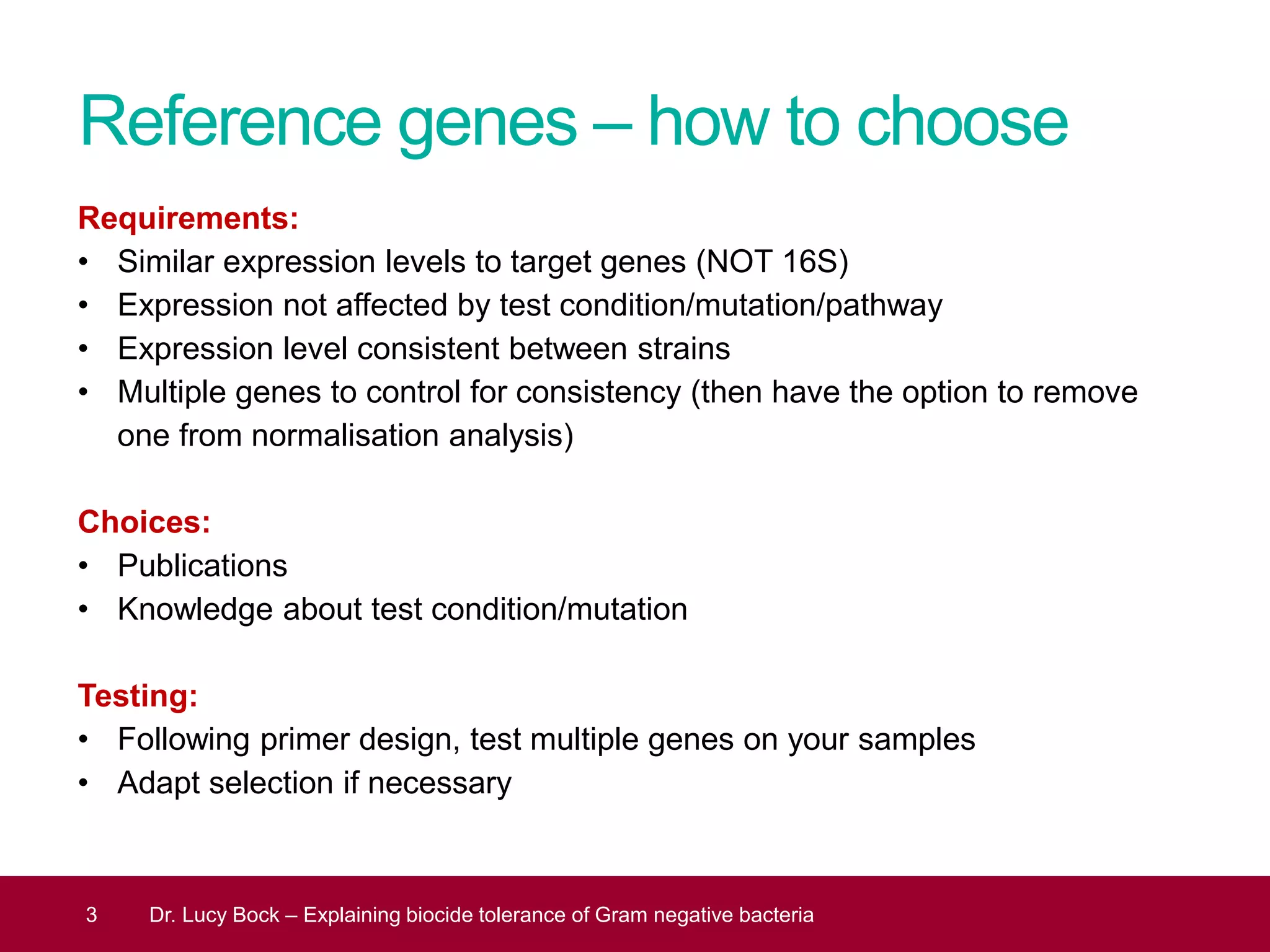
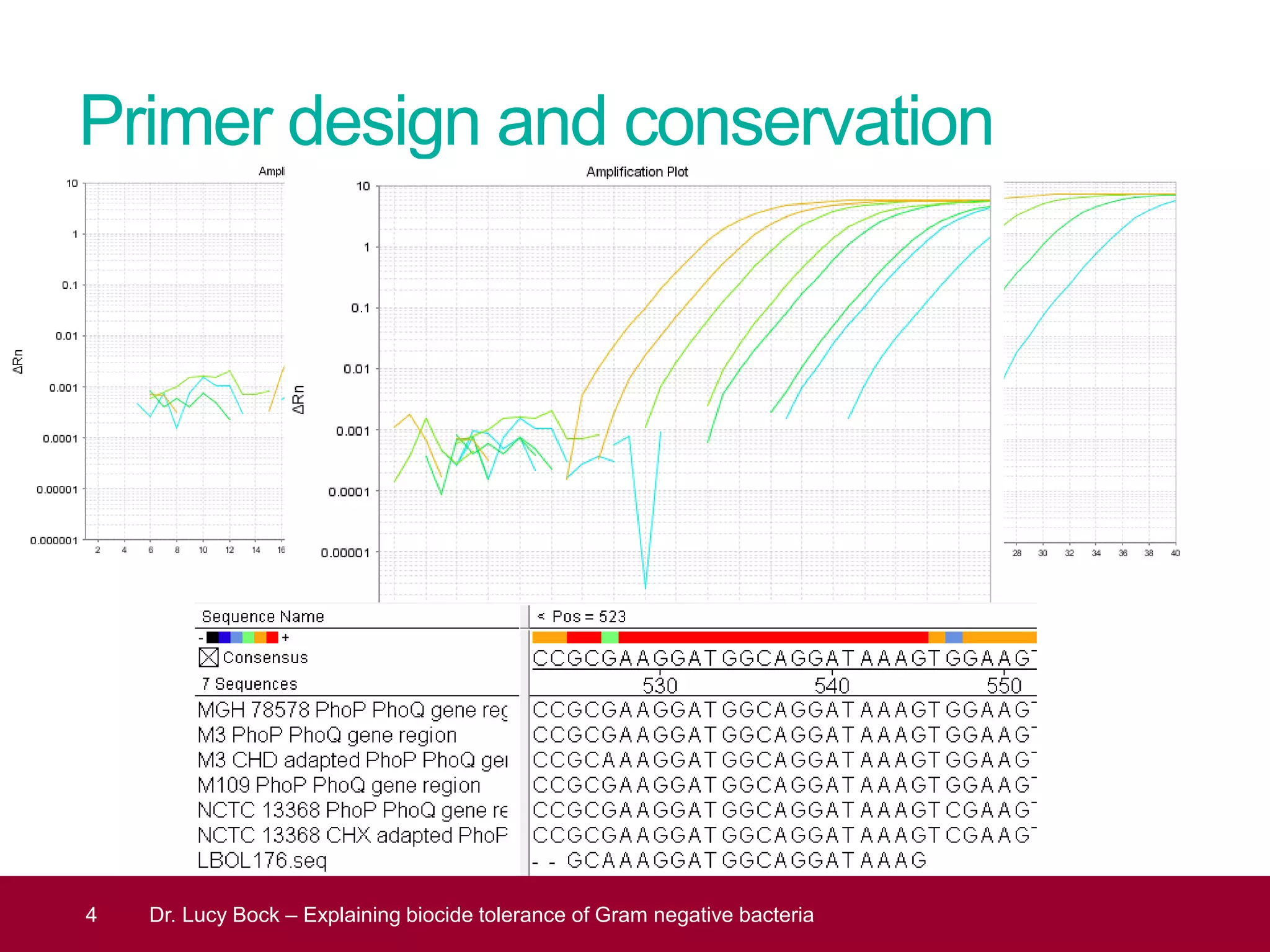
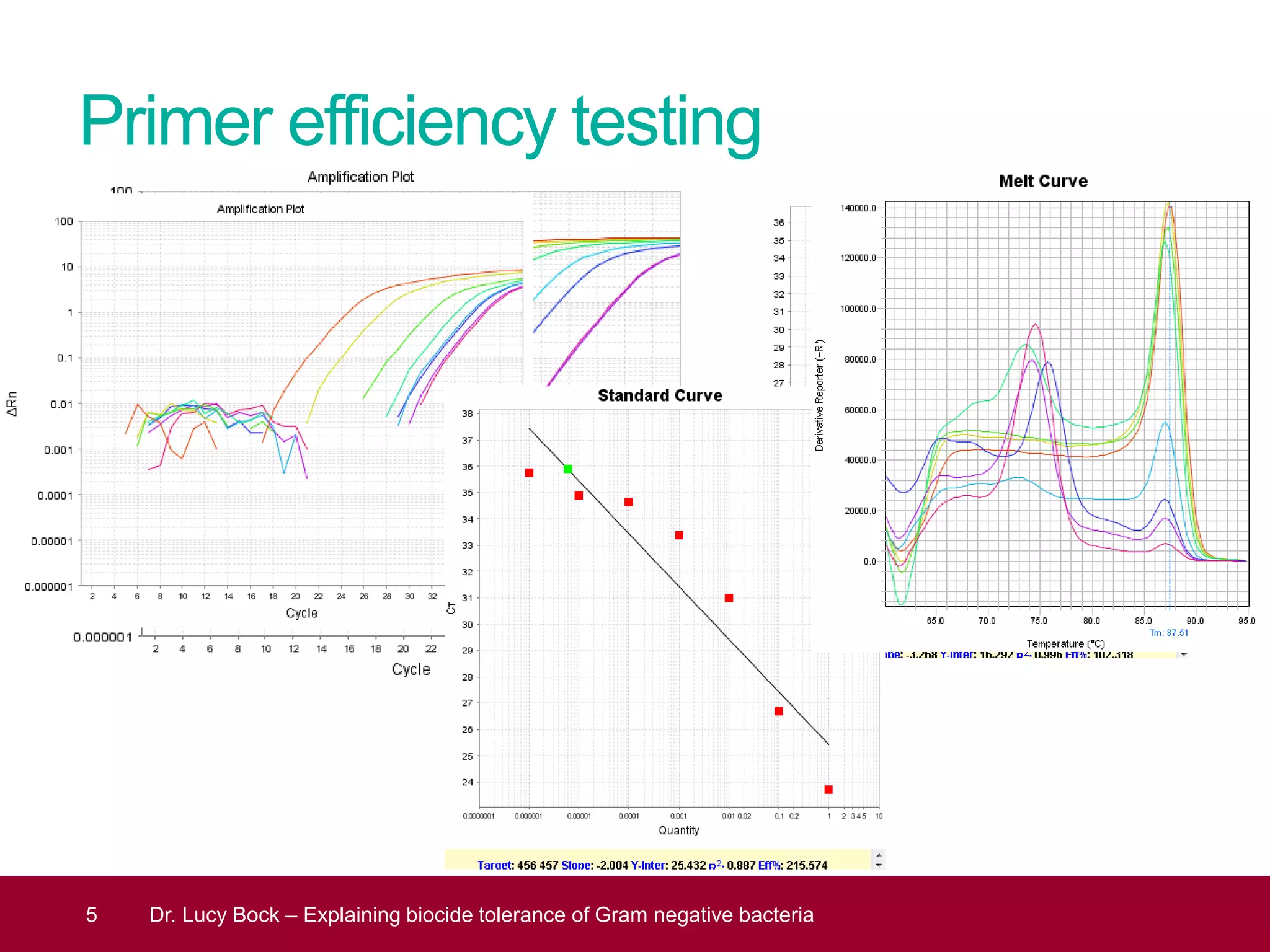
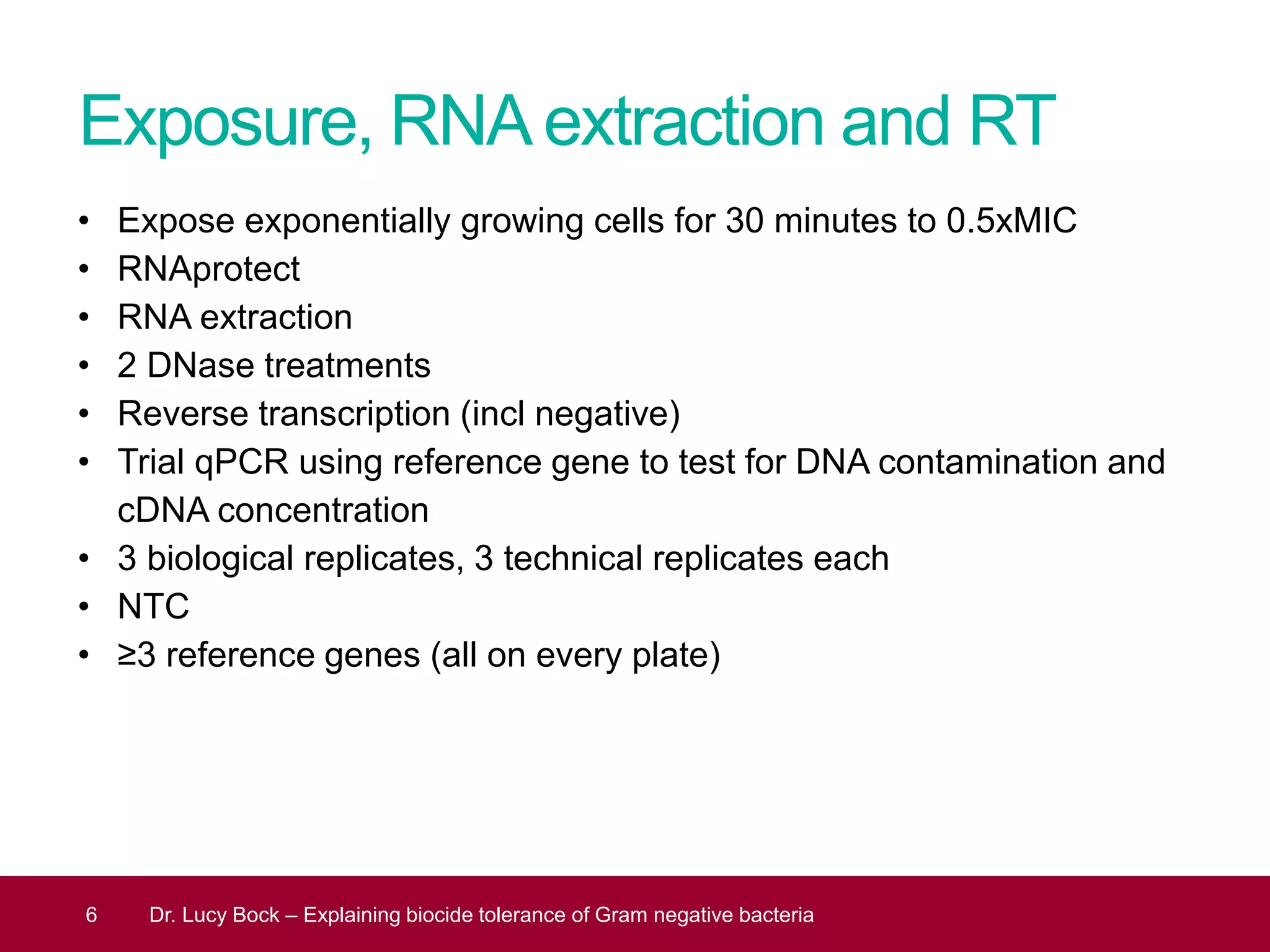
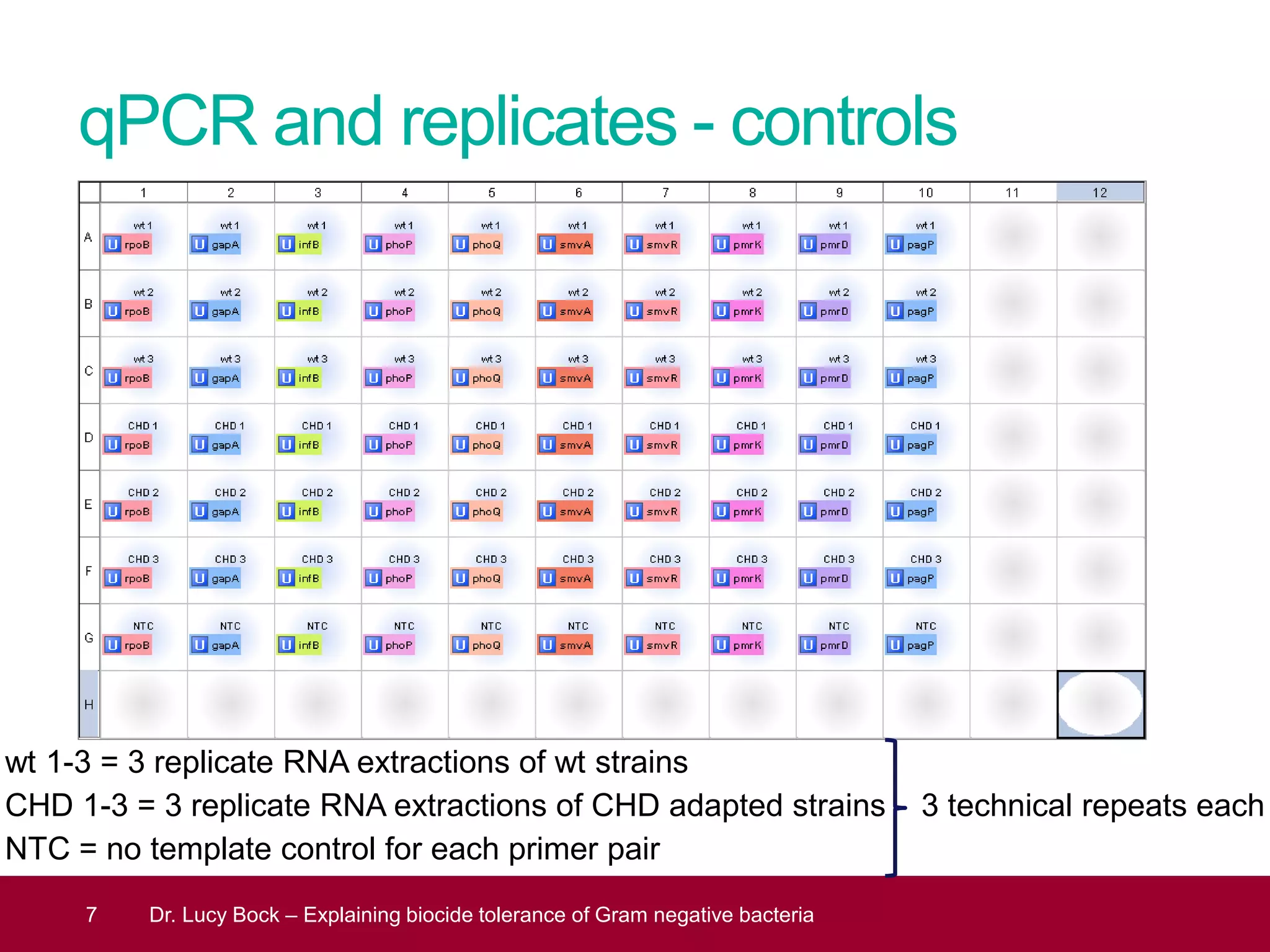
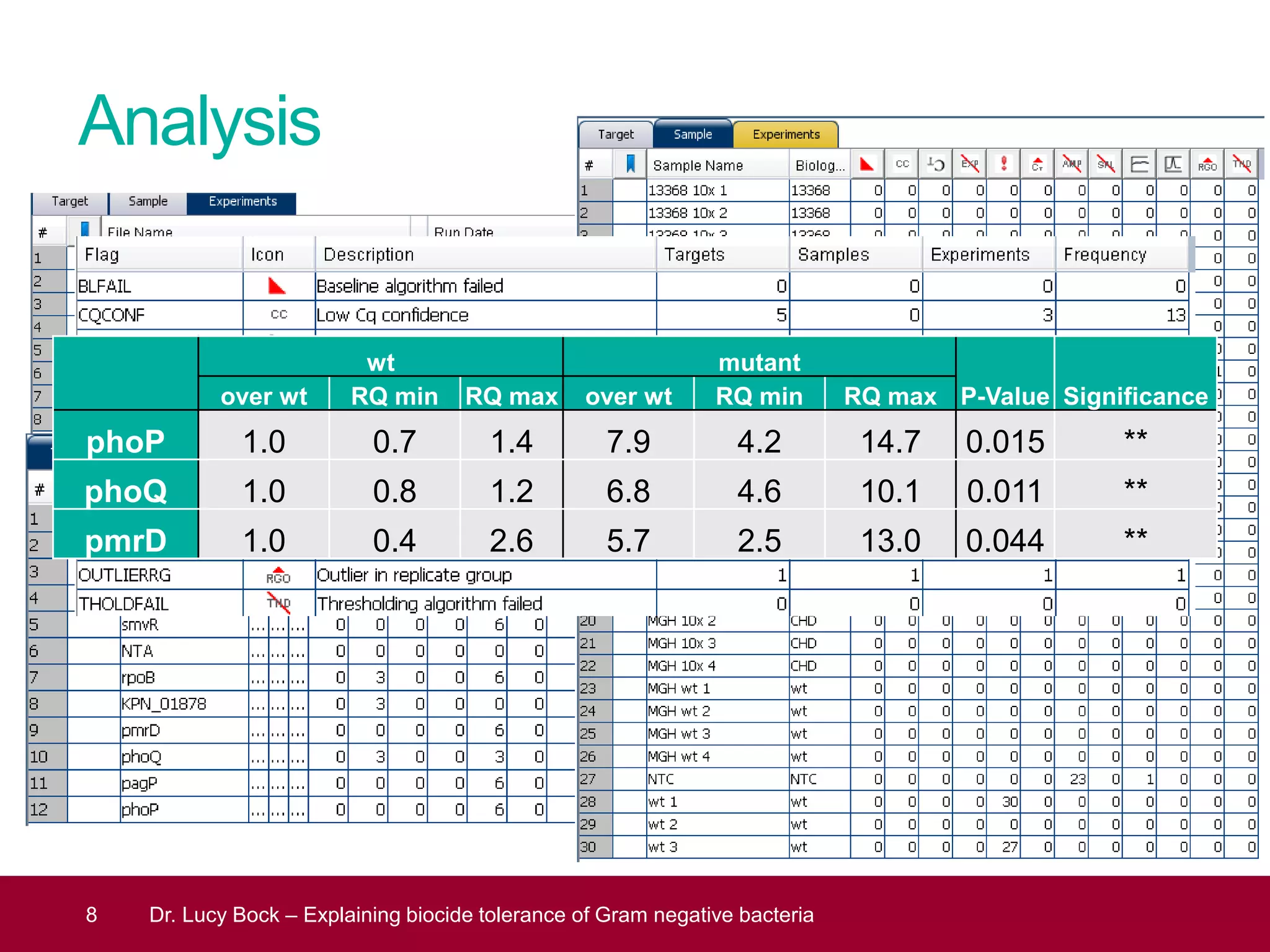
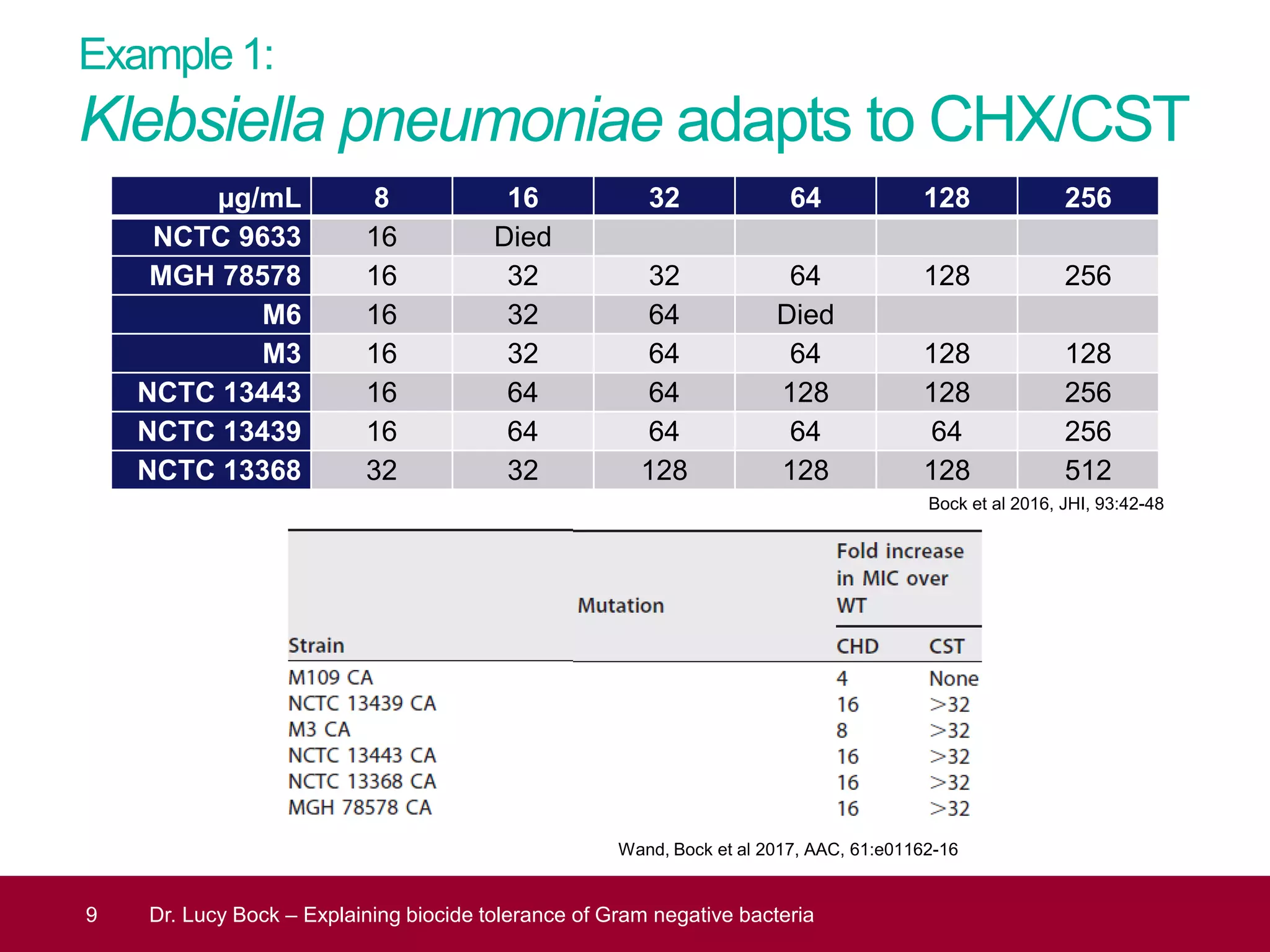
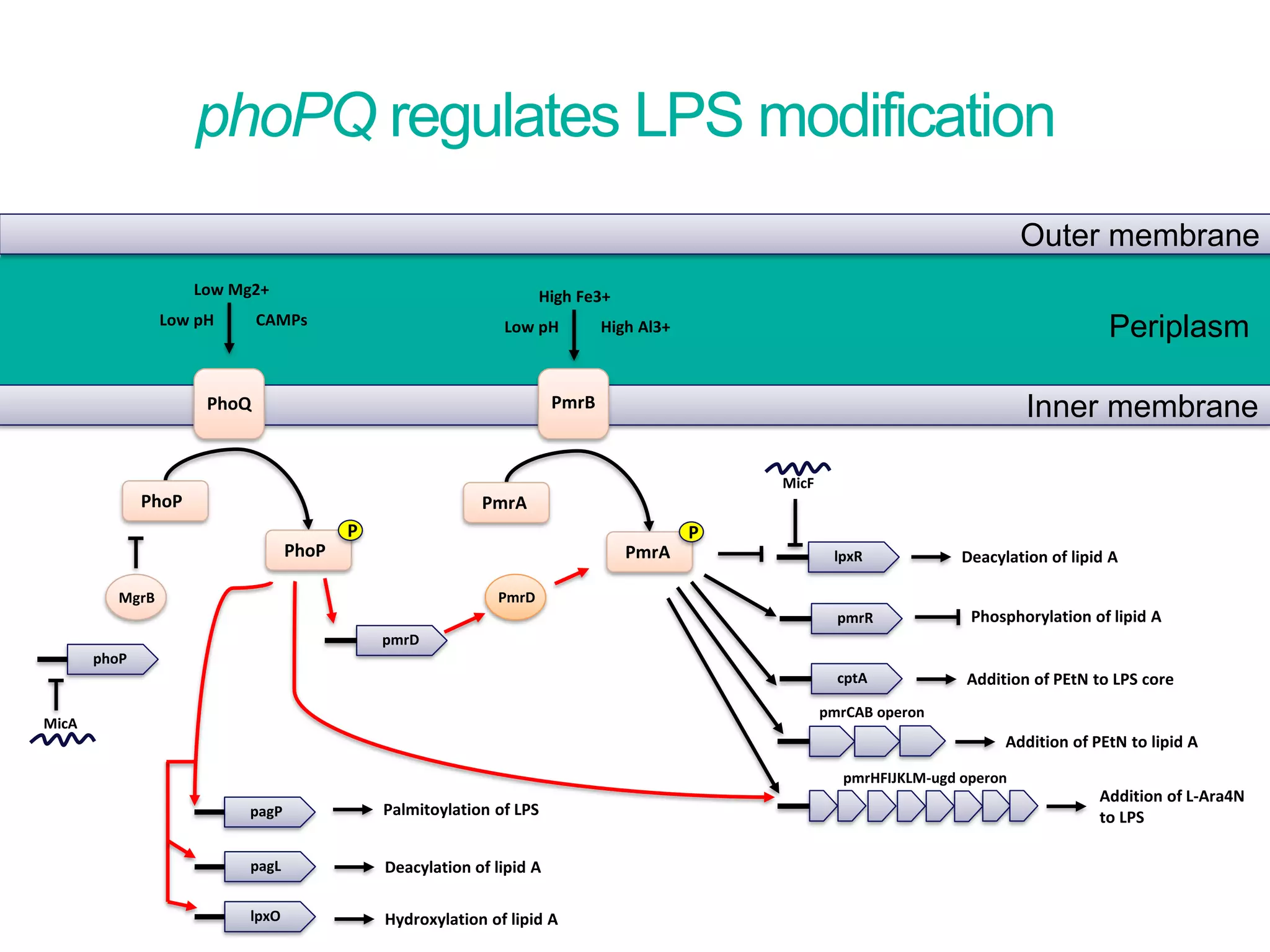
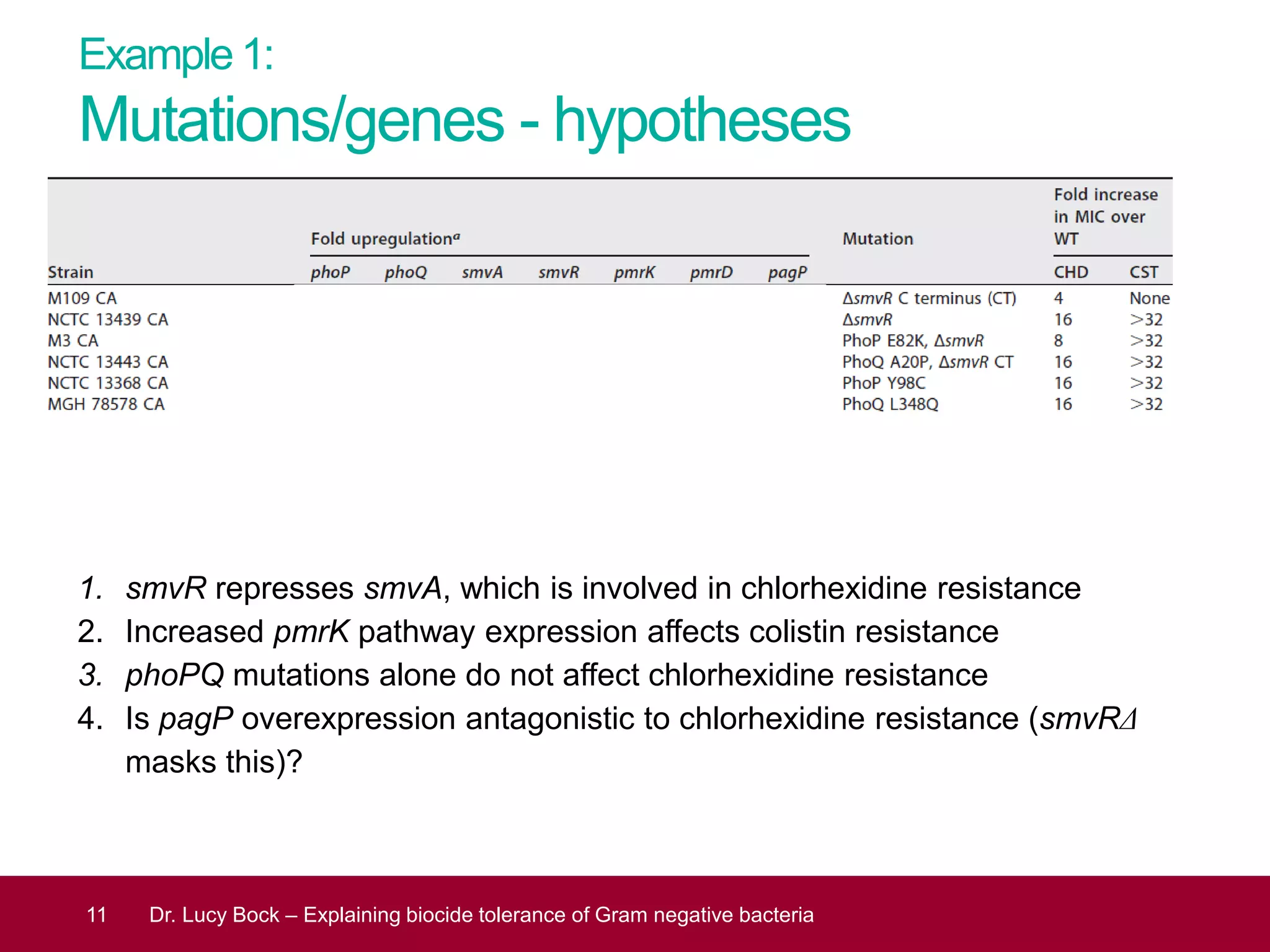
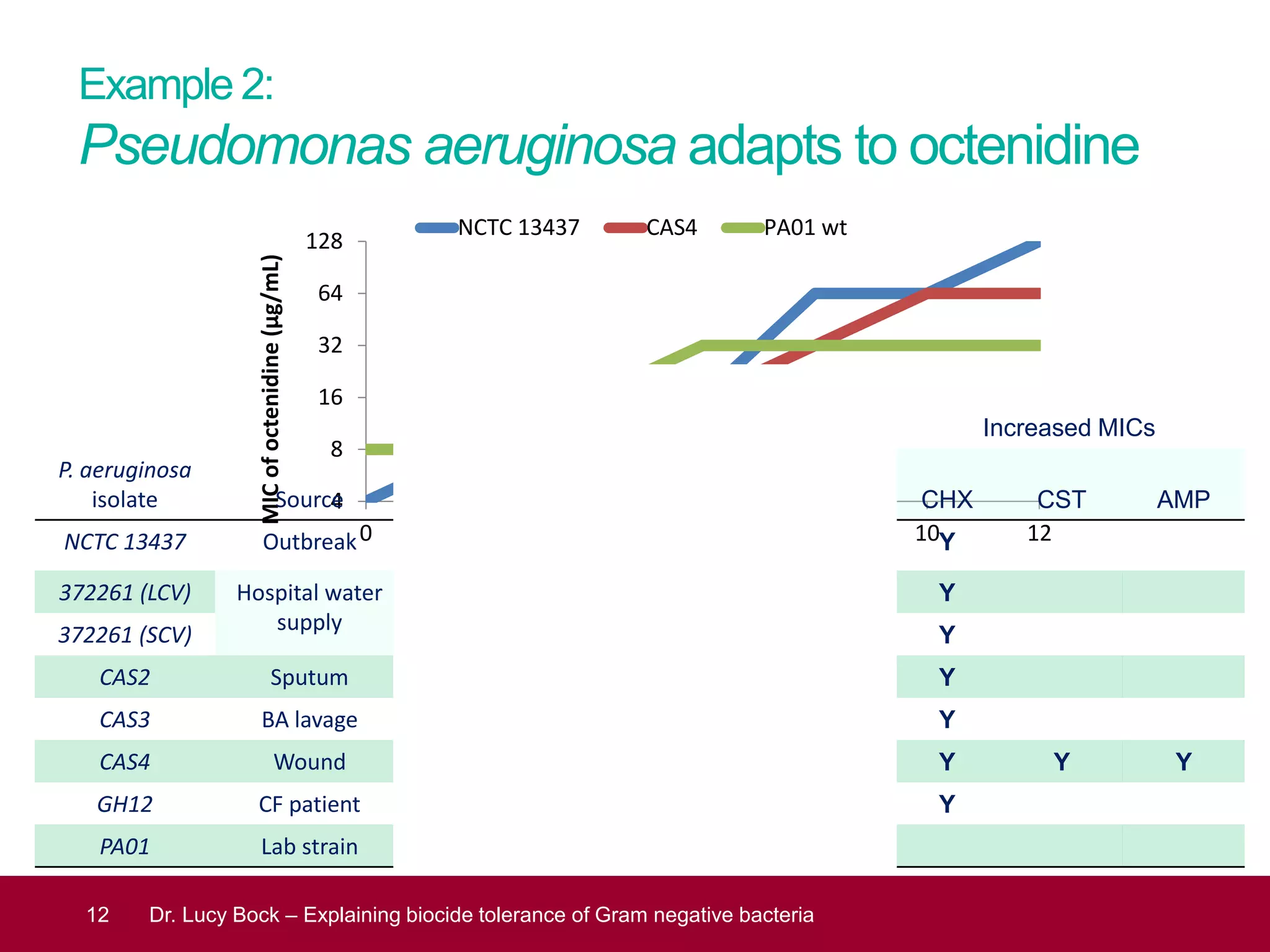
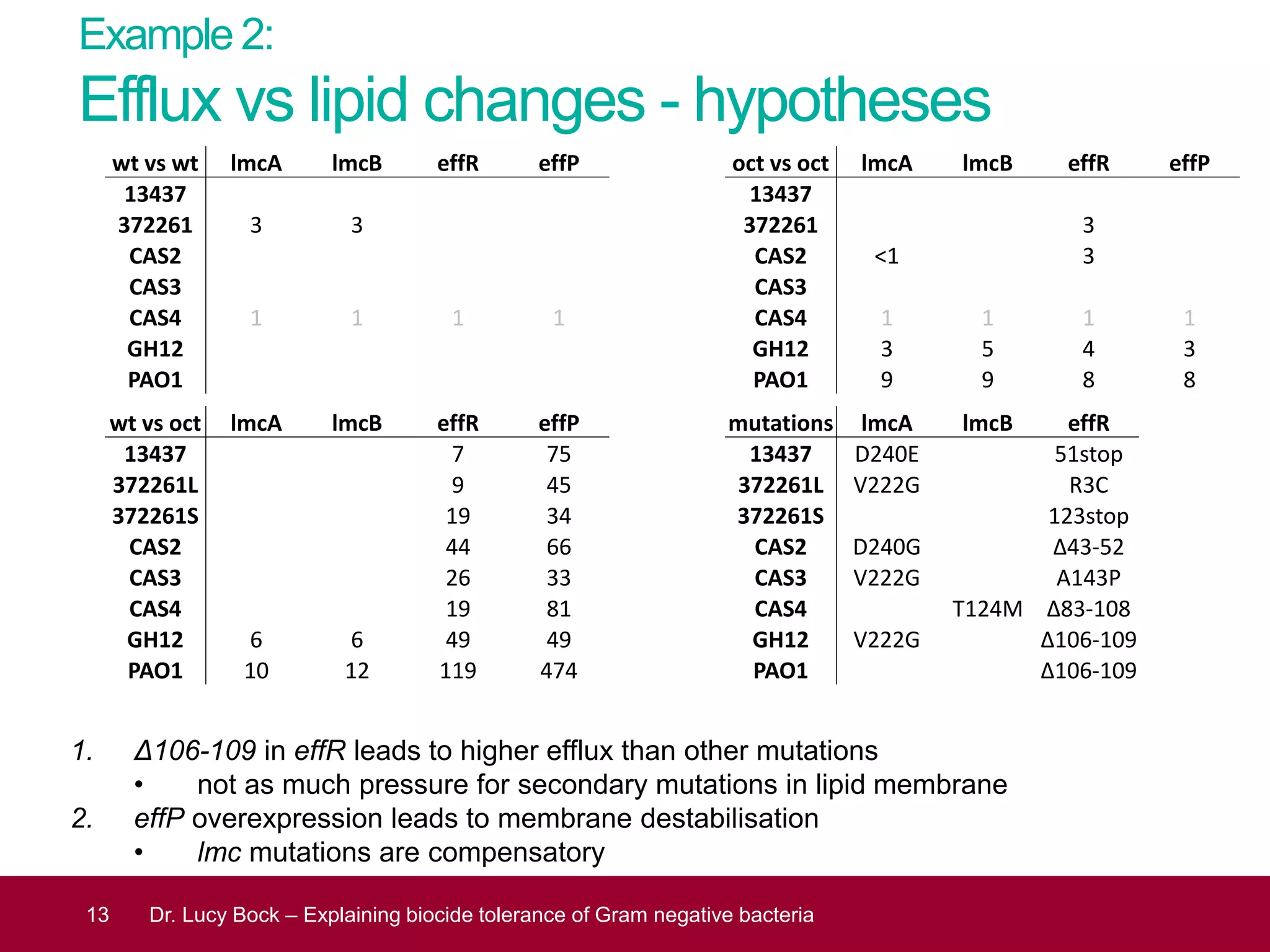
The document discusses methods to investigate biocide tolerance in Gram-negative bacteria using SYBR Green qPCR, emphasizing primer design, reference gene selection, and the significance of consistent results across different strains. It outlines experimental approaches including RNA extraction and qPCR analysis to examine the expression of target genes under various conditions, highlighting specific examples with Klebsiella pneumoniae and Pseudomonas aeruginosa. The advantages and considerations of using SYBR Green qPCR for these analyses are also covered, positioning it as a versatile tool for understanding bacterial responses to biocides.