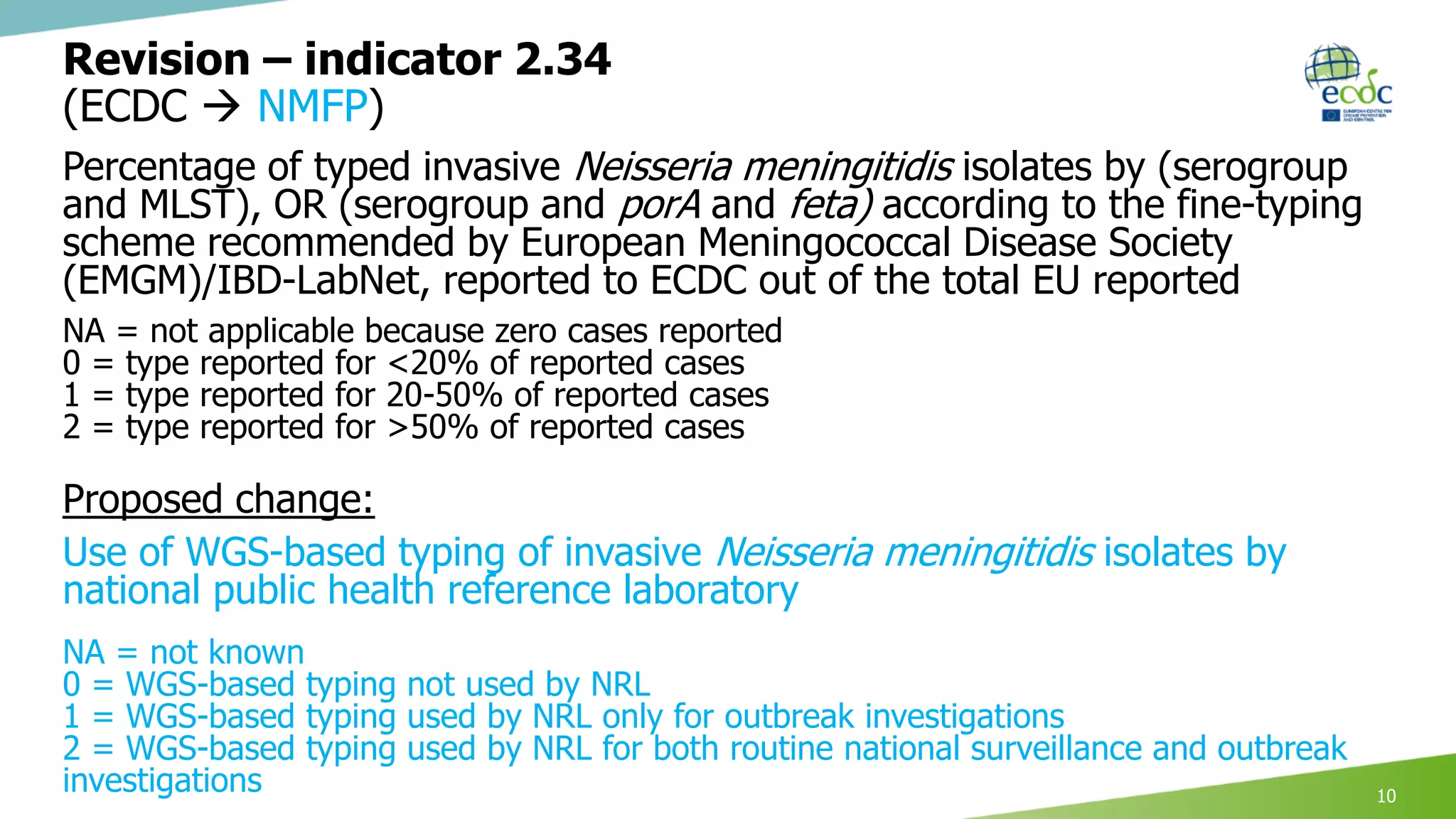
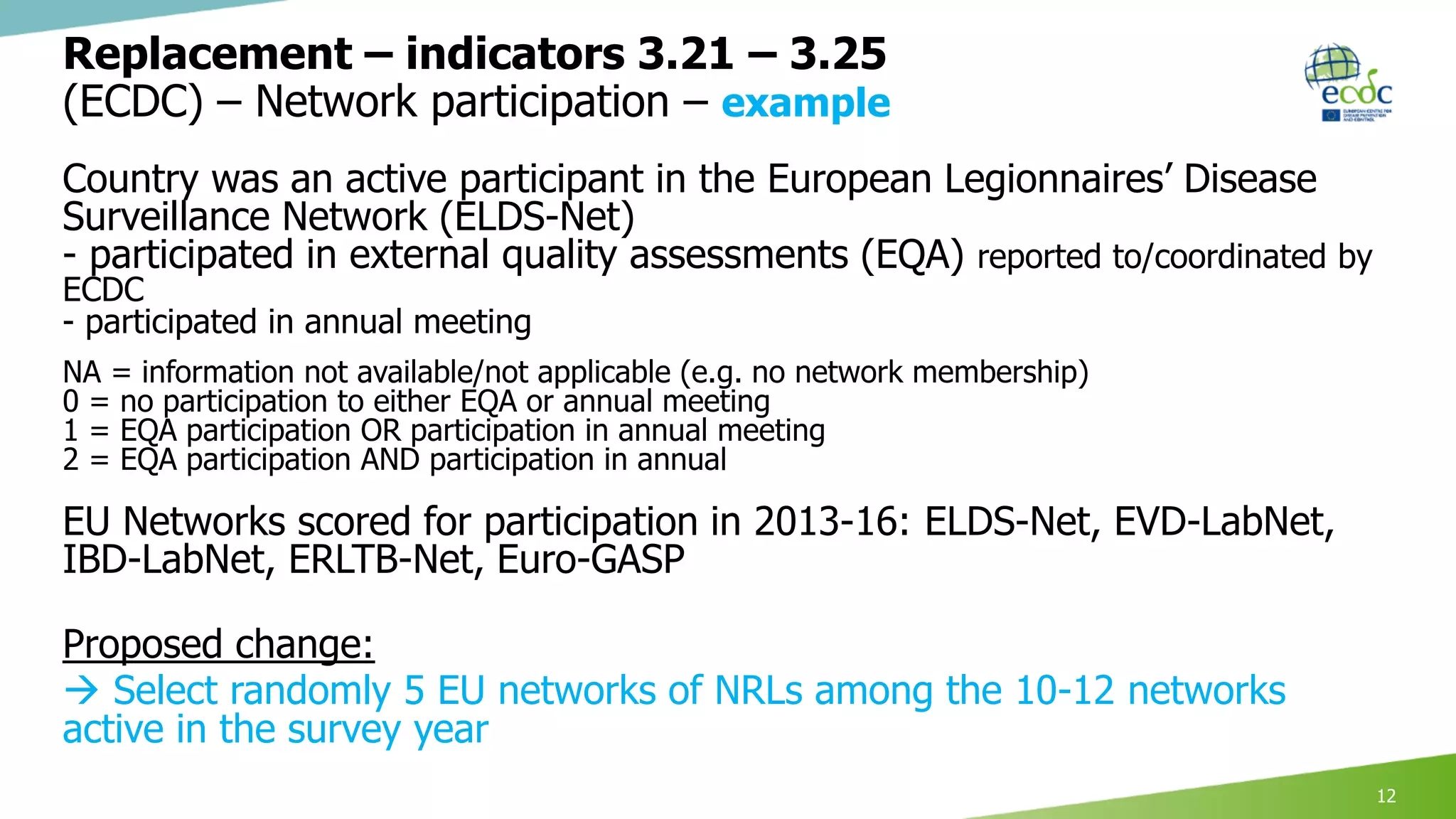
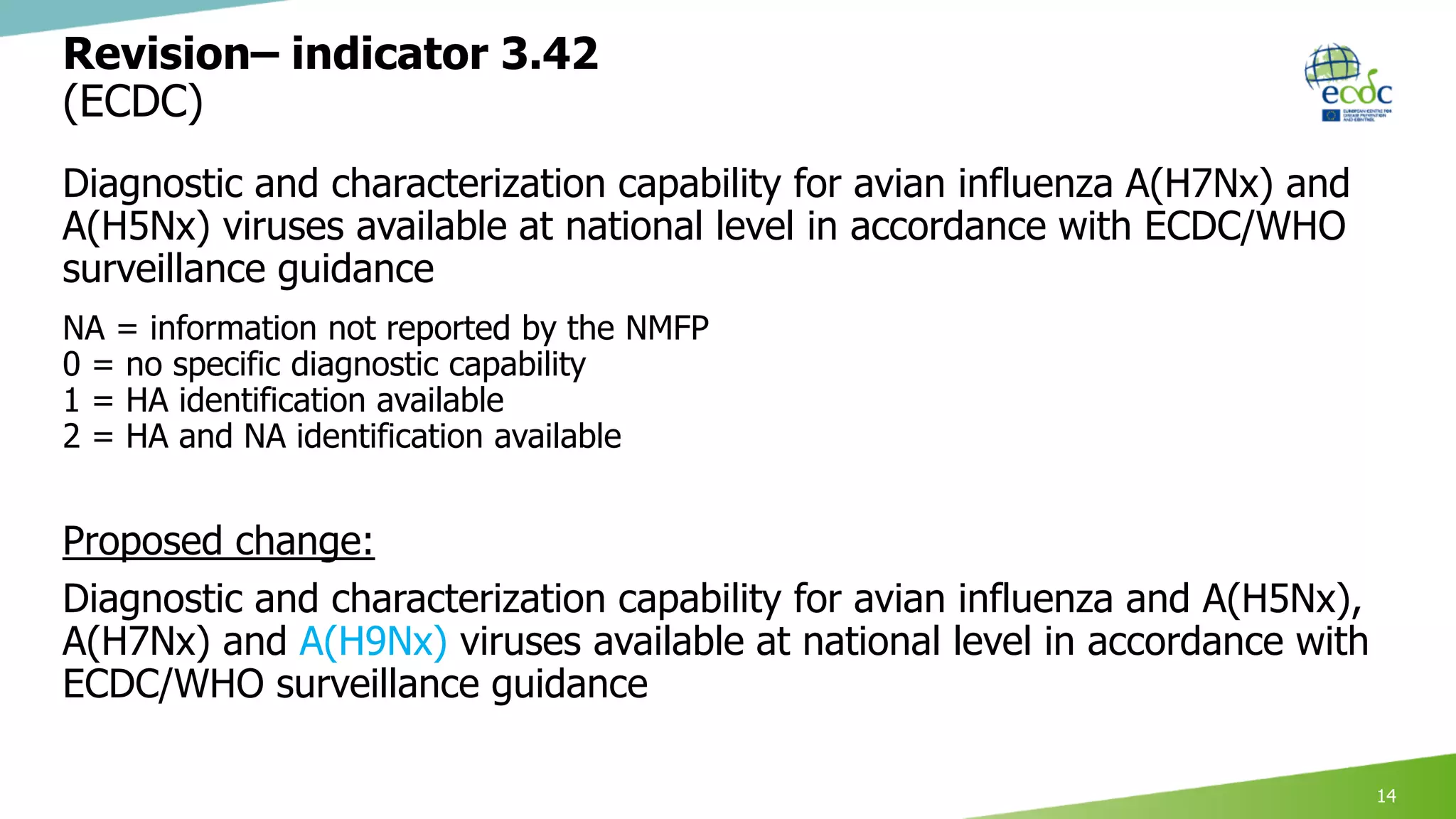
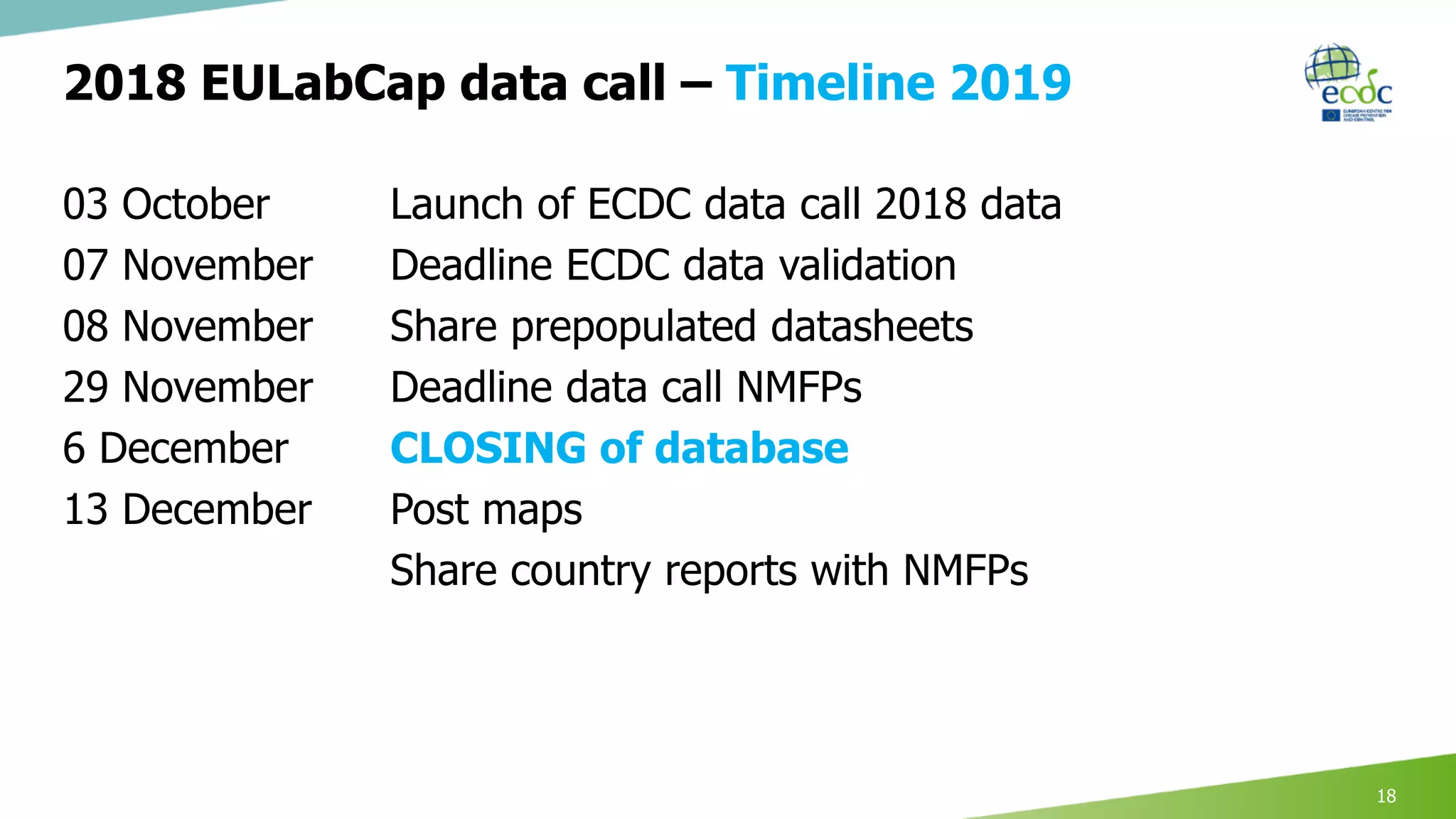
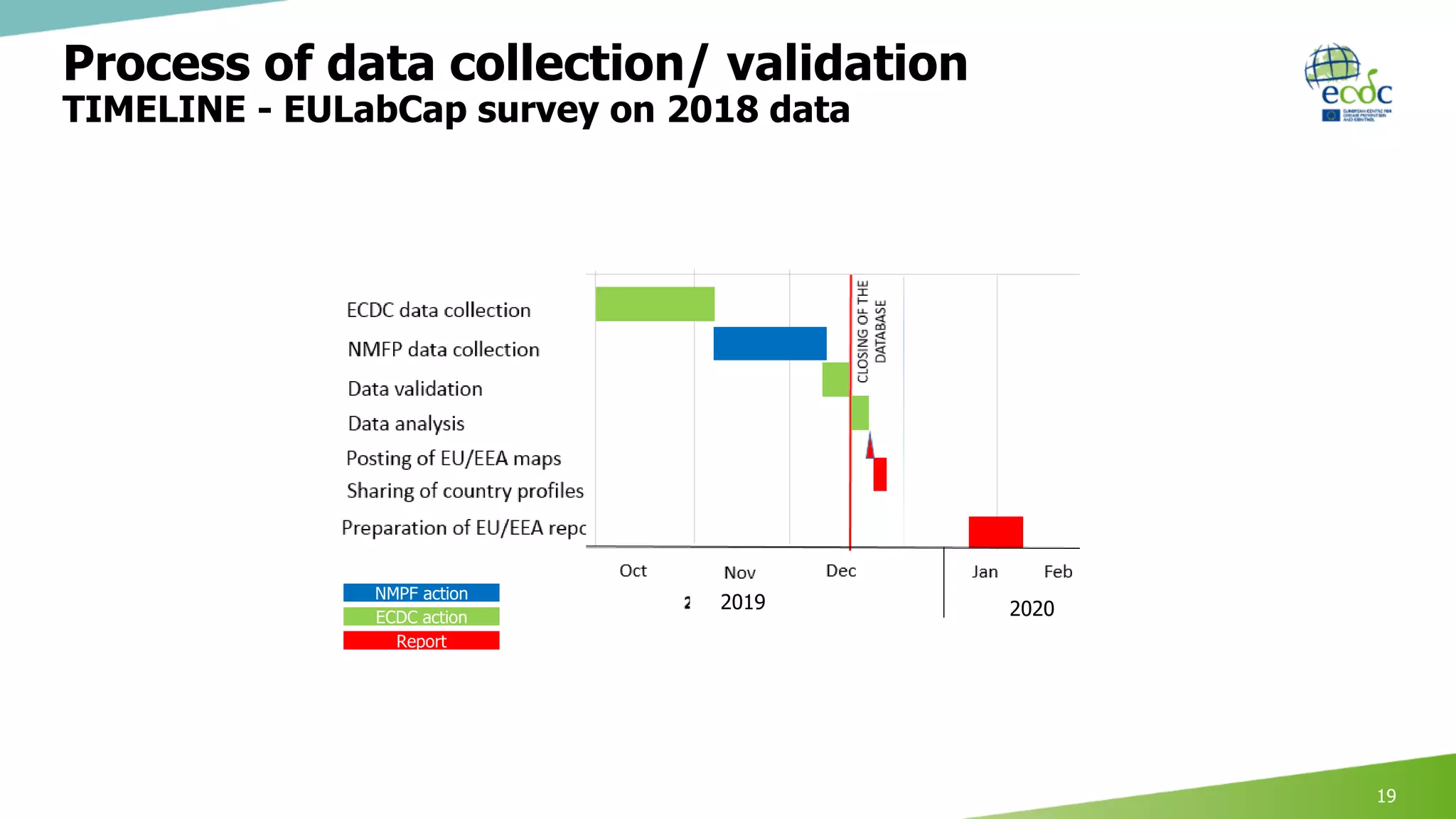
This document discusses proposed revisions to indicators and targets used in the European Union Laboratory Capability Monitoring Tool (EULabCap) survey. It provides examples of indicators being revised from using a scoring system based on participation in networks or reporting data to scoring based on absolute numbers achieved. The timeline for the 2018 EULabCap data call is also presented, including launching the call in October, a November deadline for responses from National Microbiology Focal Points, and finalizing the database and reports by December.