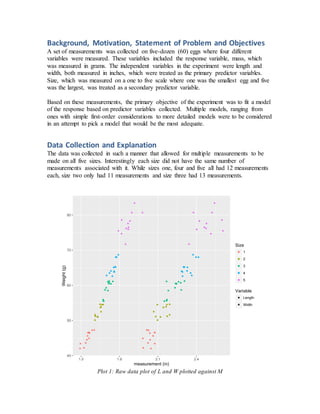
- Measurements were taken of the length, width, size, and mass of 60 eggs to develop a model predicting mass from the other variables.
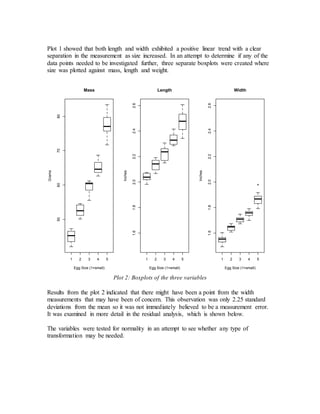
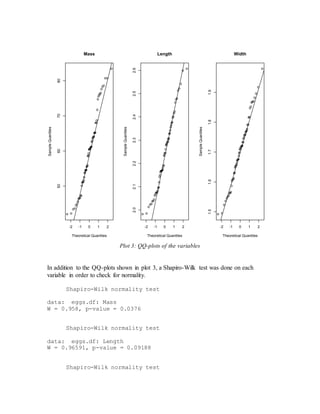
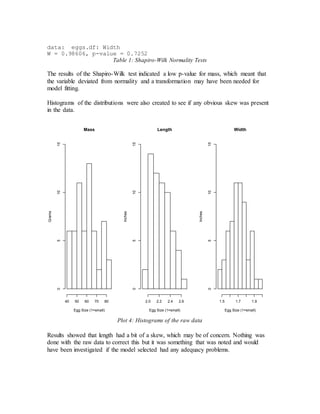
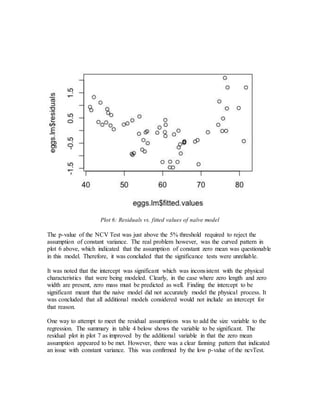
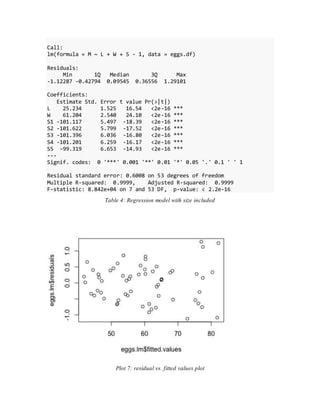
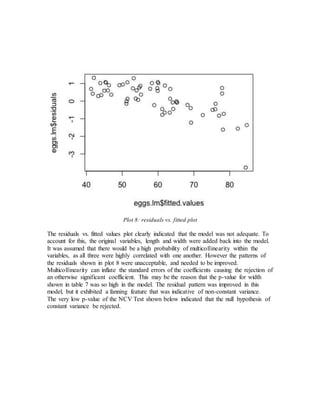
- Initial models using just length and width or adding size did not meet assumptions of constant variance.
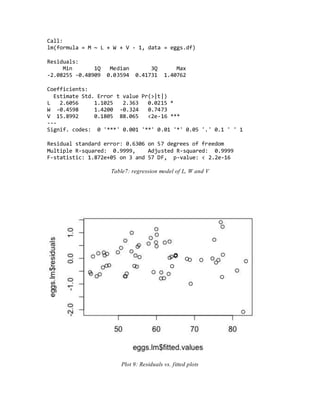
- The final adequate model included length, width, and a calculated volume variable, meeting assumptions and performing well on cross-validation.


![Statistical Analysis
The modeling effort started by constructing the naive regression model of mass as a
linear function of length and width. The summary in table 2 below shows a very high
adjusted r-squared of 0.995. All the p-values were essentially zero as well, indicating that
the intercept and both dependent variables were significant.
Call:
lm(formula = M ~ ., data = eggs.df[, 1:3])
Residuals:
Min 1Q Median 3Q Max
-1.46800 -0.49104 -0.02423 0.43205 2.09134
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -109.775 1.629 -67.38 <2e-16 ***
L 27.354 1.405 19.47 <2e-16 ***
W 63.605 2.046 31.09 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.822 on 57 degrees of freedom
Multiple R-squared: 0.9949, Adjusted R-squared: 0.9947
F-statistic: 5566 on 2 and 57 DF, p-value: < 2.2e-16
Shapiro-Wilk normality test
data: eggs.lm$residuals
W = 0.98254, p-value = 0.545
Table 2: naïve regression model
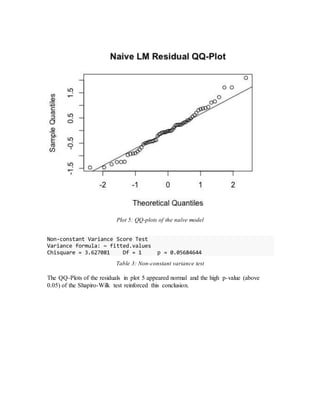
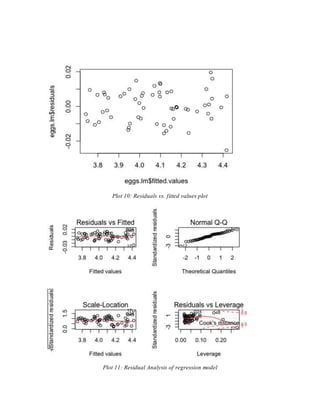
In an attempt to check model adequacy, residual analysis was done on the naïve model by
checking normality, constant variance and non-linearity.](https://image.slidesharecdn.com/f7747abc-55f4-46c1-b562-9407c928df3c-160224045209/85/eggs_project_interm-7-320.jpg)




![Appendix – R Code
library(car)
library(forecast)
library(glmnet)
eggs.df<-read.table("Eggs.txt",header = TRUE, sep = "t")
str(eggs.df)
eggs.df$S<-as.factor(eggs.df$S)
summary(scale(eggs.df[which(eggs.df$S==5),1:3]))
cor(eggs.df[,1:3])
## Question 1
units<-c("Grams","Inches","Inches")
titles<-c("Mass", "Length", "Width")
quartz()
par(mfrow=c(1,3))
boxplot(eggs.df[,1]~eggs.df$S, main=titles[1],
xlab="Egg Size (1=small)",ylab=units[1])
for (i in 2:3){
boxplot(eggs.df[,i]~eggs.df$S, main=titles[i],
ylim=c(min(eggs.df[,2:3]),max(eggs.df[,2:3])),
xlab="Egg Size (1=small)",ylab=units[i])
}
## or
quartz()
par(mfrow=c(1,3))
for (i in 1:3){
boxplot(eggs.df[,i]~eggs.df$S, main=titles[i],
xlab="Egg Size (1=small)",ylab=units[i])
}](https://image.slidesharecdn.com/f7747abc-55f4-46c1-b562-9407c928df3c-160224045209/85/eggs_project_interm-18-320.jpg)
![## Histograms
quartz()
par(mfrow=c(1,3))
for (i in 1:3){
hist(eggs.df[,i], main=titles[i], ylim = c(0,15),
xlab="Egg Size (1=small)",ylab=units[i])
# lines(density(eggs.df[,i]), col="blue", lwd=2)
}
# Notice Skew of Length
## Check Normality
quartz()
par(mfrow=c(1,3))
for (i in 1:3){
qqnorm(eggs.df[,i],main=titles[i])
qqline(eggs.df[,i])
}
shapiro.test(eggs.df[,1])
shapiro.test(eggs.df[,2])
shapiro.test(eggs.df[,3])
# Appears Normal
## Question 2.d without S
eggs.lm<-lm(M~., data=eggs.df[,1:3])
summary(eggs.lm)](https://image.slidesharecdn.com/f7747abc-55f4-46c1-b562-9407c928df3c-160224045209/85/eggs_project_interm-19-320.jpg)

