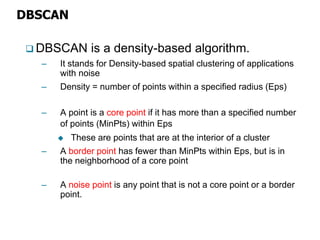
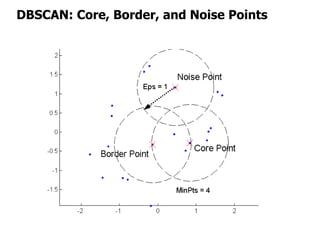
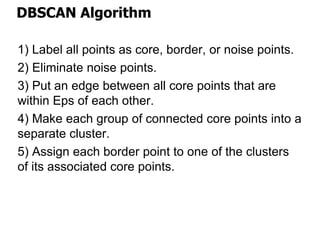
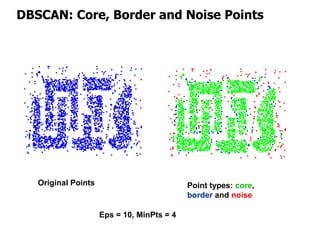
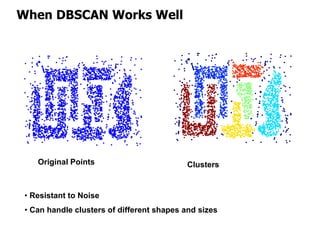
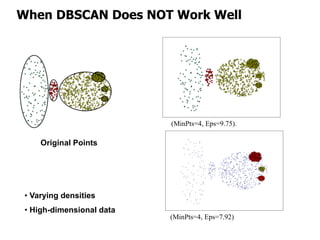
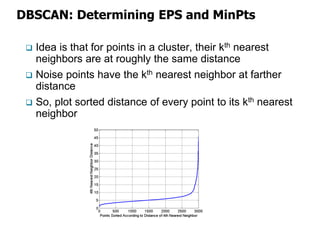
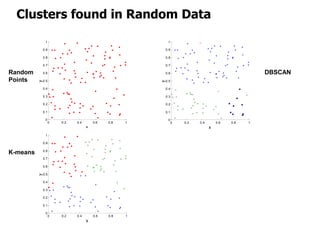
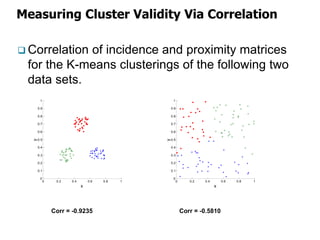
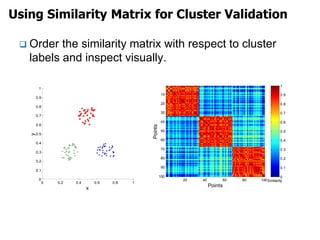
El documento describe el algoritmo de agrupamiento DBSCAN, que se basa en la densidad para identificar puntos centrales, de borde y de ruido en un conjunto de datos. Se discuten técnicas para la validez de los clusters, incluyendo índices externos e internos, y se muestran métodos para evaluar la calidad de los grupos obtenidos. También se abordan aspectos como la validación mediante matrices de proximidad y correlación para medir la efectividad del agrupamiento.