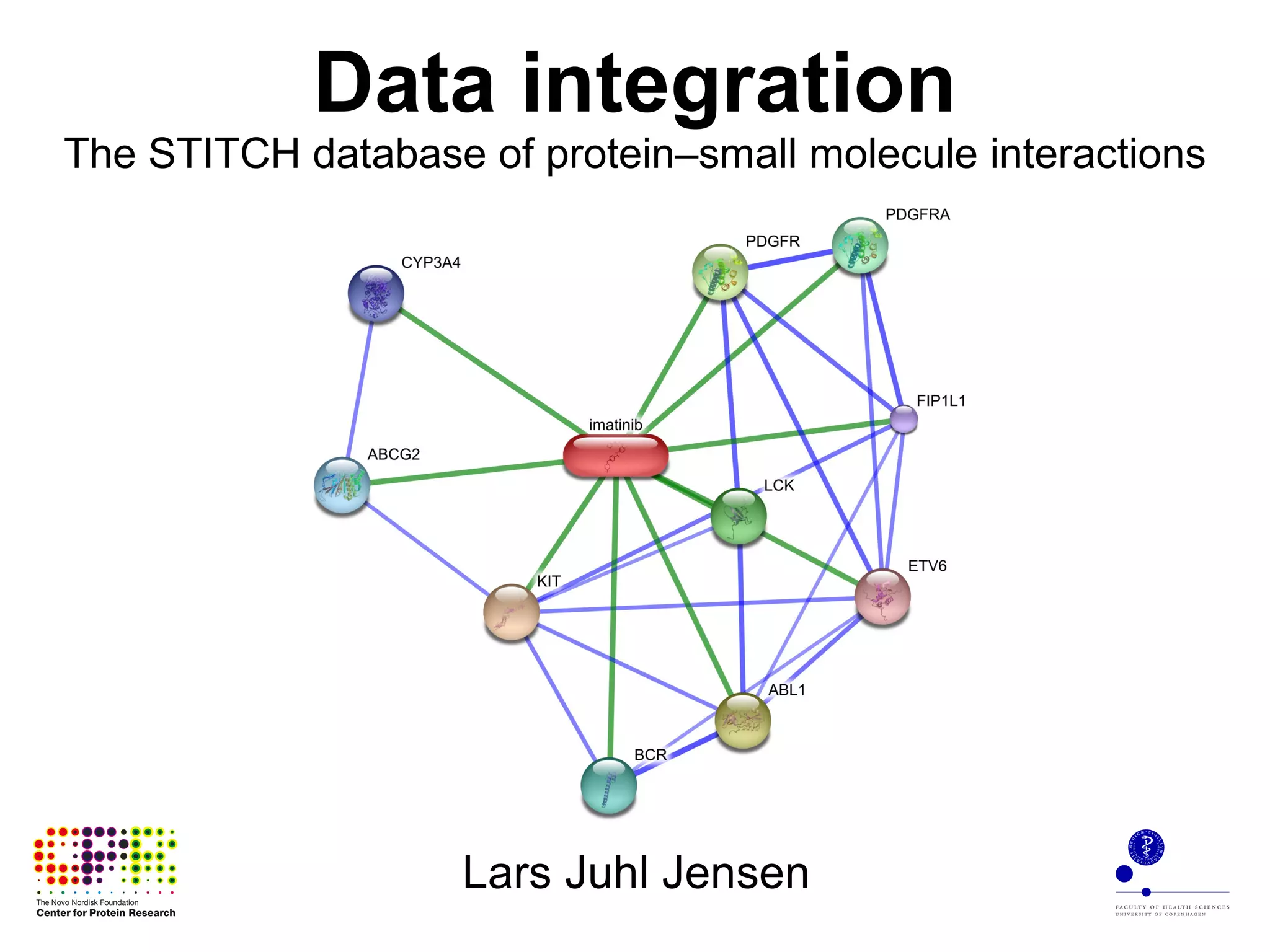
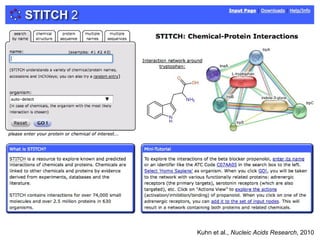
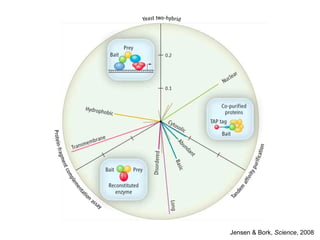
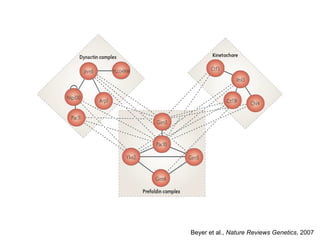
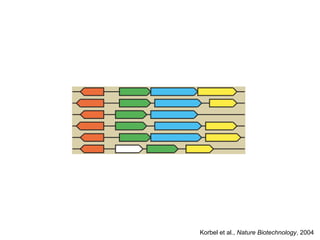
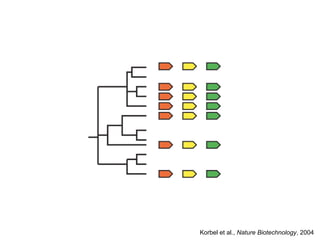
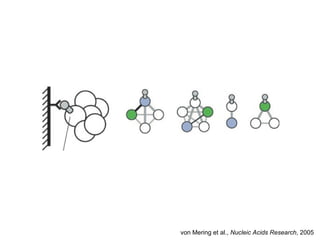
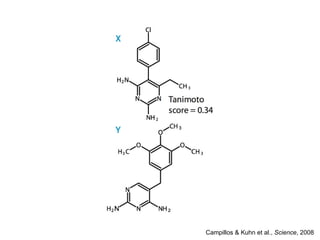
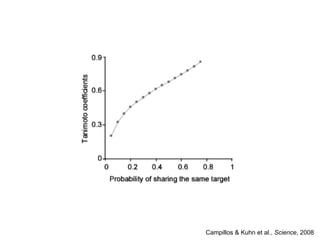
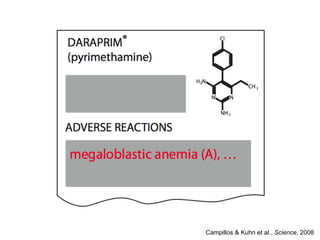
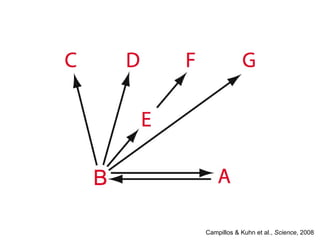
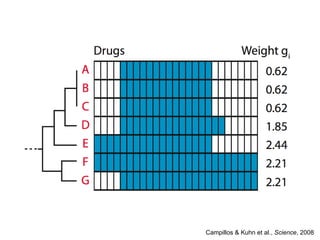
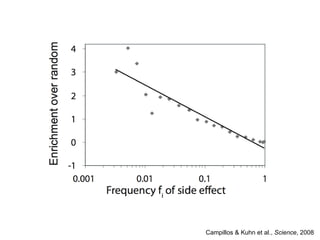
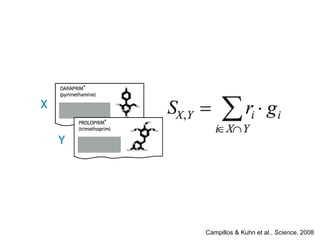
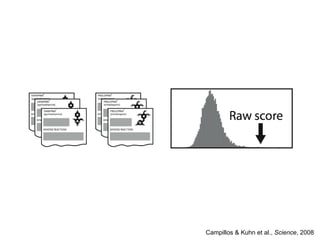
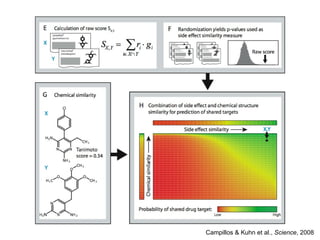
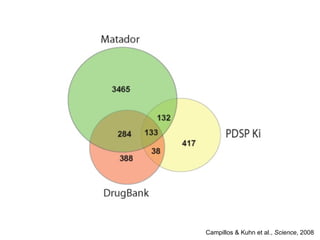
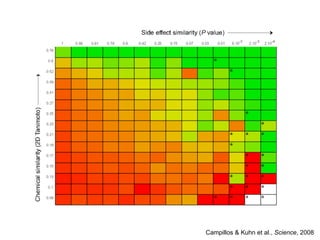
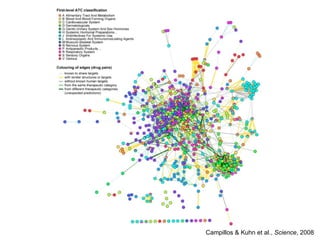
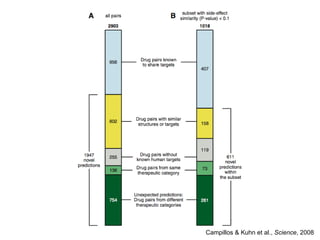
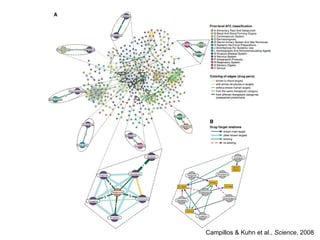
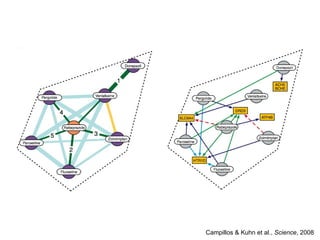
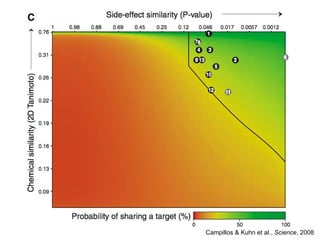
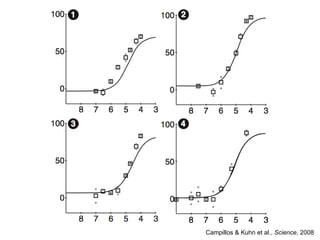
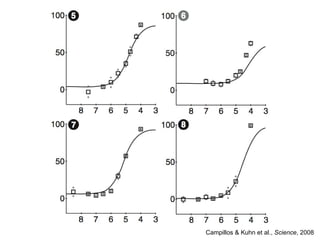
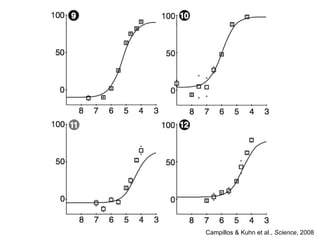
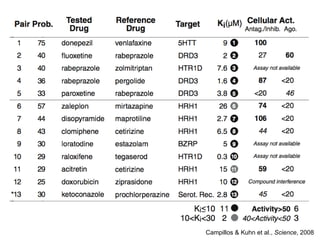
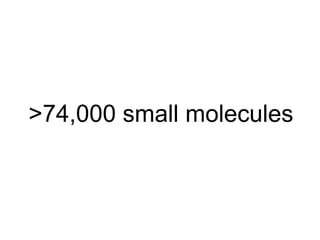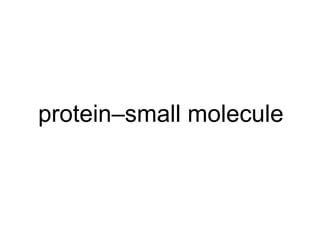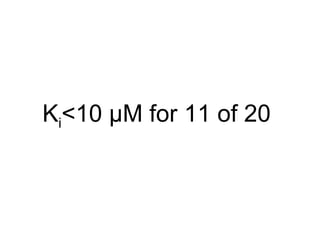The document discusses data integration and summarizes the STITCH database, which integrates protein-small molecule interactions from several sources. It also summarizes a method for predicting novel drug targets using side effect information by analyzing similarities in side effect profiles between existing drugs. Predictions were tested in vitro and in cell assays, with promising results. The document acknowledges contributions from researchers involved in developing STITCH and the side effect prediction method.