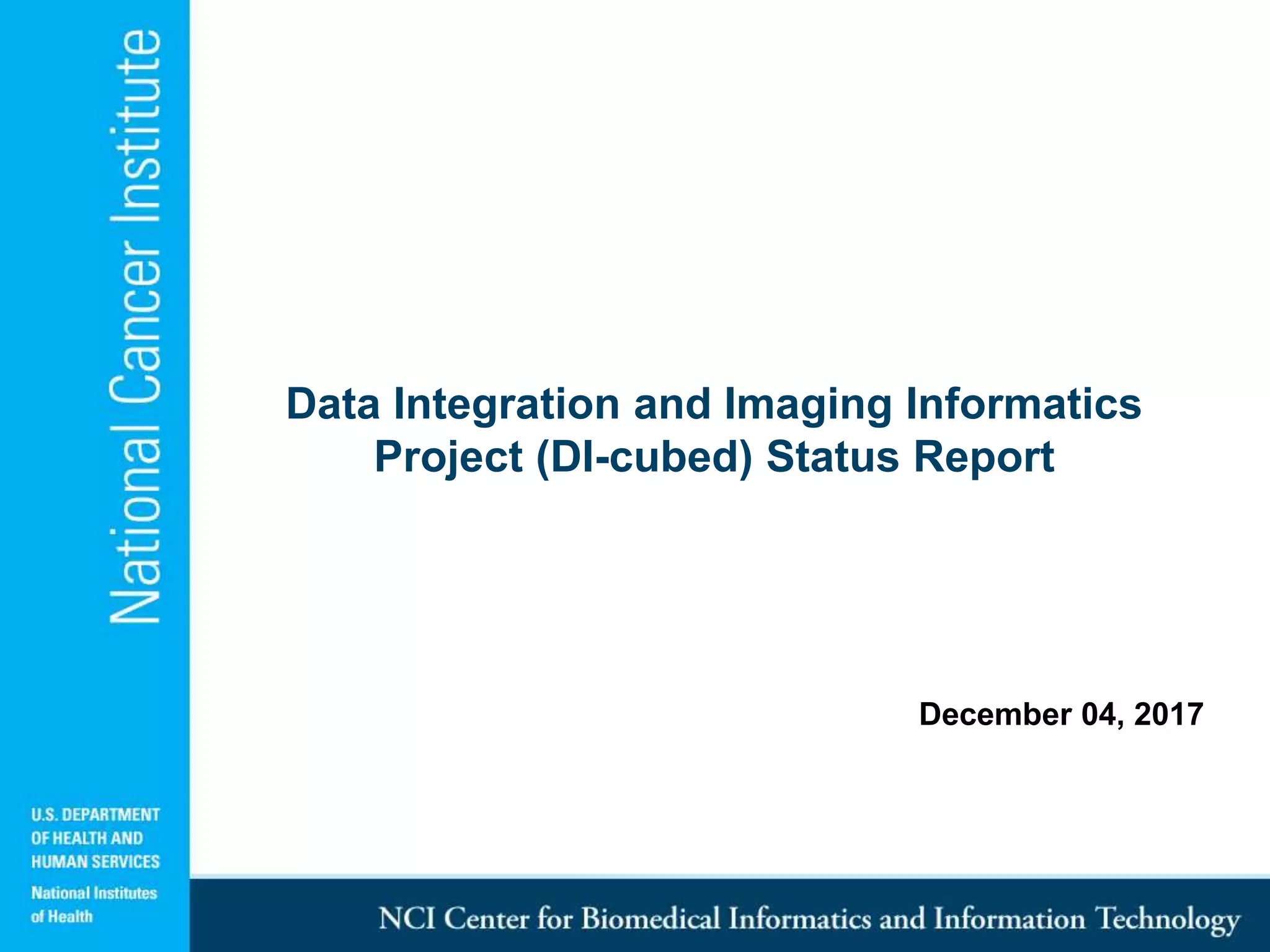
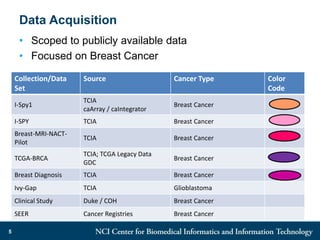
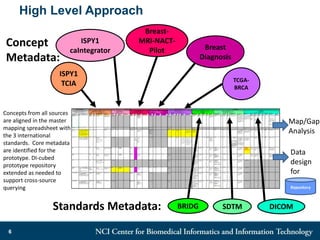
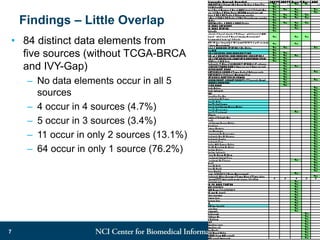
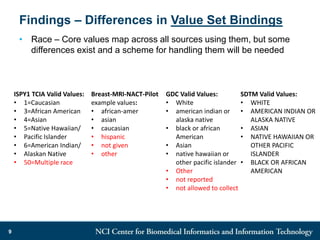
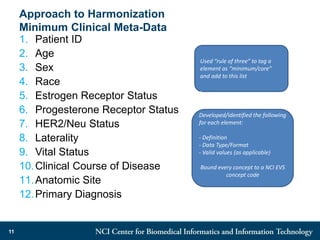
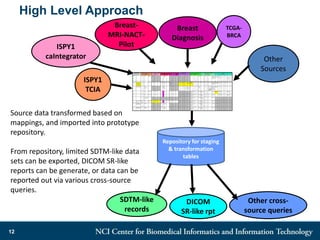
The DI-CUBED project aims to facilitate interoperability between clinical data and imaging standards, using frameworks like BRIDG, CDISC, and DICOM. A prototype is being developed to allow querying across disparate datasets while highlighting differences in semantic consistency. Recommendations include standardizing clinical data sets to improve efficiency in data collection and enhance integration with larger research initiatives.