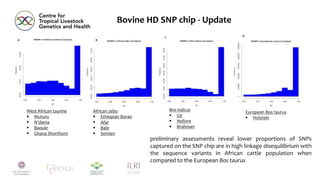
This document summarizes an ongoing project to create a genomic reference resource for African cattle (GRRFAC). The project aims to sequence 600-700 samples from 40 African cattle breeds. So far, 71 samples have been sequenced with 225 more planned by end of 2019. Analysis of sequences shows 38 million SNPs identified across samples. The document also summarizes assessments of ascertainment bias in commercial SNP chips for African cattle, finding lower linkage between chip SNPs and sequence variants in African populations compared to European cattle. Finally, genome-wide scans have identified signatures of selection in African cattle related to heat tolerance, trypanotolerance, and other adaptive traits.