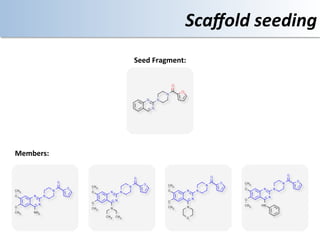
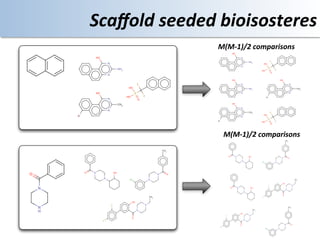
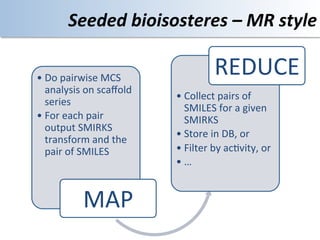
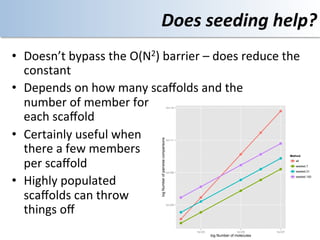
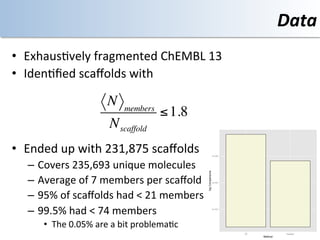
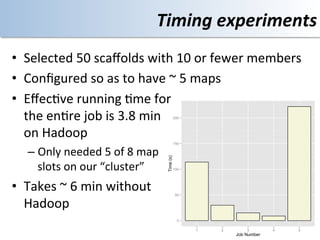
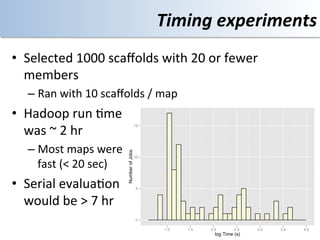
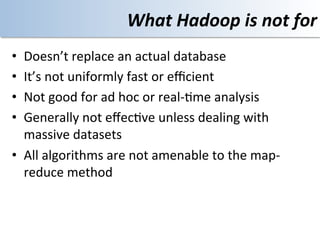
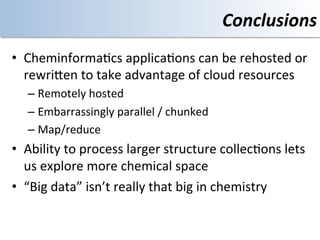
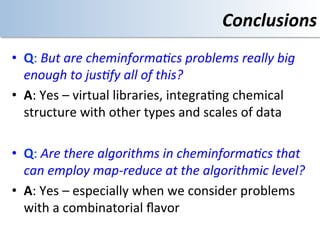
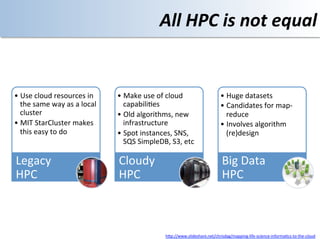
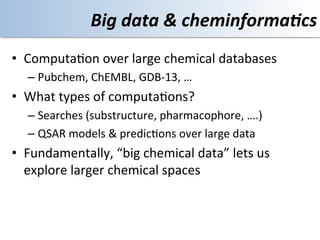
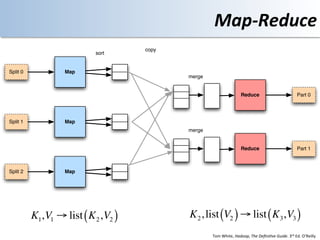
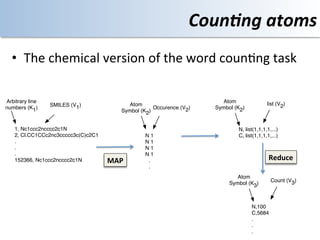
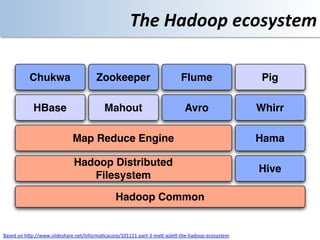
This document discusses using cloud computing resources for cheminformatics applications. It describes how Hadoop and MapReduce can be used to perform large-scale parallel computations on chemical data and databases. Specific examples discussed include counting atoms in large datasets using MapReduce and performing substructure searches using SMARTS queries on Hadoop. The document also compares different approaches to programming Hadoop applications and how Pig Latin can simplify writing cheminformatics jobs for Hadoop.

![Simplifying
Hadoop
applica4ons
package gov.nih.ncgc.hadoop;
public void map(LongWritable key, Text value, OutputCollector<Text, IntWritable> output, Reporter
import chemaxon.formats.MolFormatException;
reporter) throws IOException {
import chemaxon.formats.MolImporter;
• Raw
Hadoop
Molecule mol = MolImporter.importMol(value.toString());
import chemaxon.license.LicenseManager;
matches.set(mol.getName());
import chemaxon.license.LicenseProcessingException;
search.setTarget(mol);
import chemaxon.sss.search.MolSearch;
try {
import chemaxon.sss.search.SearchException;
if (search.isMatching()) {
import chemaxon.struc.Molecule;
output.collect(matches, one);
import org.apache.hadoop.conf.Configuration;
} else {
programs
can
import org.apache.hadoop.conf.Configured;
output.collect(matches, zero);
import org.apache.hadoop.filecache.DistributedCache;
}
import org.apache.hadoop.fs.Path;
} catch (SearchException e) {
import org.apache.hadoop.io.IntWritable;
}
import org.apache.hadoop.io.LongWritable;
}
import org.apache.hadoop.io.Text;
}
import org.apache.hadoop.mapred.FileInputFormat;
be
tedious
to
import org.apache.hadoop.mapred.FileOutputFormat;
public static class SmartsMatchReducer extends MapReduceBase implements Reducer<Text,
import org.apache.hadoop.mapred.JobClient;
IntWritable, Text, IntWritable> {
import org.apache.hadoop.mapred.JobConf;
private IntWritable result = new IntWritable();
import org.apache.hadoop.mapred.MapReduceBase;
import org.apache.hadoop.mapred.Mapper;
public void reduce(Text key,
import org.apache.hadoop.mapred.OutputCollector;
Iterator<IntWritable> values,
import org.apache.hadoop.mapred.Reducer;
OutputCollector<Text, IntWritable> output,
write
import org.apache.hadoop.mapred.Reporter;
Reporter reporter) throws IOException {
import org.apache.hadoop.mapred.TextInputFormat;
while (values.hasNext()) {
import org.apache.hadoop.mapred.TextOutputFormat;
if (values.next().compareTo(one) == 0) {
import org.apache.hadoop.util.Tool;
result.set(1);
import org.apache.hadoop.util.ToolRunner;
output.collect(key, result);
}
import java.io.BufferedReader;
}
import java.io.FileReader;
}
import java.io.IOException;
}
import java.util.Iterator;
public int run(String[] args) throws Exception {
/**
JobConf jobConf = new JobConf(getConf(), HeavyAtomCount.class);
* SMARTS searching over a set of files using Hadoop.
jobConf.setJobName("smartsSearch");
*
* @author Rajarshi Guha
jobConf.setOutputKeyClass(Text.class);
*/
jobConf.setOutputValueClass(IntWritable.class);
public class SmartsSearch extends Configured implements Tool {
private final static IntWritable one = new IntWritable(1);
jobConf.setMapperClass(MoleculeMapper.class);
private final static IntWritable zero = new IntWritable(0);
jobConf.setCombinerClass(SmartsMatchReducer.class);
jobConf.setReducerClass(SmartsMatchReducer.class);
public static class MoleculeMapper extends MapReduceBase implements Mapper<LongWritable, Text,
Text, IntWritable> {
jobConf.setInputFormat(TextInputFormat.class);
private String pattern = null;
jobConf.setOutputFormat(TextOutputFormat.class);
private MolSearch search;
jobConf.setNumMapTasks(5);
public void configure(JobConf job) {
if (args.length != 4) {
try {
System.err.println("Usage: ss <in> <out> <pattern> <license file>");
Path[] licFiles = DistributedCache.getLocalCacheFiles(job);
System.exit(2);
BufferedReader reader = new BufferedReader(new FileReader(licFiles[0].toString()));
}
StringBuilder license = new StringBuilder();
String line;
FileInputFormat.setInputPaths(jobConf, new Path(args[0]));
while ((line = reader.readLine()) != null) license.append(line);
FileOutputFormat.setOutputPath(jobConf, new Path(args[1]));
reader.close();
jobConf.setStrings("pattern", args[2]);
LicenseManager.setLicense(license.toString());
} catch (IOException e) {
// make the license file available vis dist cache
} catch (LicenseProcessingException e) {
DistributedCache.addCacheFile(new Path(args[3]).toUri(), jobConf);
}
JobClient.runJob(jobConf);
pattern = job.getStrings("pattern")[0];
return 0;
search = new MolSearch();
}
try {
Molecule queryMol = MolImporter.importMol(pattern, "smarts");
public static void main(String[] args) throws Exception {
search.setQuery(queryMol);
} catch (MolFormatException e) {
int res = ToolRunner.run(new Configuration(), new SmartsSearch(), args);
}
}
SMARTS
based
}
}
final static IntWritable one = new IntWritable(1);
Text matches = new Text();
substructure
search](https://image.slidesharecdn.com/guha-cloud-ul-120926095626-phpapp02/85/Cloudy-with-a-Touch-of-Cheminformatics-9-320.jpg)