Recommended
PPT
Chapter :Tour of cell ,structure and function of parts
PPT
PPTX
PPT
06atourofthecell-130311053323-phpapp01 [Autoguardado].ppt
PPT
PDF
PPT
PPTX
06 Lecture BIOL 1010-30 Gillette College
PPT
Eukaryotic cell Prokaryotic cell.ppt
PPT
PDF
6_Cell Structure and Detailed Overview.pdf
PPT
Power pt on cell structure
PPT
Sel Campbelll 8 untuk olimpiade SMA .ppt
PDF
PPT
PPT
PPT
PDF
Cellular Structure and Processes AP Biology
PPTX
7. The Cell - hoe cells work and organelles.pptx
PPT
PPT
PDF
PPTX
Cell Structure and Function
PPTX
PPTX
Mod 04 Tour of the Cell Presentation.pptx
PPT
PPT
PPTX
L1 Introduction to cells.pptx
PPT
Cahpter 04-Carbon Introductor course Camp
PPT
Cahpter 03-Water Wasser Introductor course
More Related Content
PPT
Chapter :Tour of cell ,structure and function of parts
PPT
PPTX
PPT
06atourofthecell-130311053323-phpapp01 [Autoguardado].ppt
PPT
PDF
PPT
PPTX
06 Lecture BIOL 1010-30 Gillette College
Similar to Chapter 6 Introductory course Biology Camp
PPT
Eukaryotic cell Prokaryotic cell.ppt
PPT
PDF
6_Cell Structure and Detailed Overview.pdf
PPT
Power pt on cell structure
PPT
Sel Campbelll 8 untuk olimpiade SMA .ppt
PDF
PPT
PPT
PPT
PDF
Cellular Structure and Processes AP Biology
PPTX
7. The Cell - hoe cells work and organelles.pptx
PPT
PPT
PDF
PPTX
Cell Structure and Function
PPTX
PPTX
Mod 04 Tour of the Cell Presentation.pptx
PPT
PPT
PPTX
L1 Introduction to cells.pptx
More from abomajid13
PPT
Cahpter 04-Carbon Introductor course Camp
PPT
Cahpter 03-Water Wasser Introductor course
PPT
Chapter 51 introductory biology course Camp
PPT
Chapter 52 introductory biology course Camp
PPT
Chapter 5 introductory Biology Course Camp
PPTX
Chapter 2 Introductory course Biology Camp
PPTX
Chapter 1 Introductory course Biology Camp
Recently uploaded
PDF
Solid Waste Management (SWM) _2026003456
PDF
Age of exploration - ppt presentation_EAPT
PPT
Electric Fields and capacitance AQA A-level Physics
PDF
Detection of an NH3 Absorption Band at 2.2μm on Europa
PPT
Lesson 1- Earthquake and Type of Faults.ppt
PPTX
1. BEHAVIOURAL SCIENCE An Introduction for Medical Students.pptx
PPTX
History_Taking_and_Physical_Examination_of_Cardiovascular_System.pptx
PDF
How common are oxygenic photosynthesis and large coal deposits on exoplanets?
PPTX
Mount and repair a fishing net for fishi
PPTX
ELectromagnitism.pptx ELectromagnitism.p
PPTX
OECD 204 (Acute Dermal Toxicity) .pptx
PDF
Virtualization 1: Virtual Machine, Hypervisor (Virtual Machine Monitor)
PDF
Polymer [ बहुलक ] Chemistry Notes PDF - Irfanullah Mehar - JJ Sir Chemistry.pdf
PPTX
Spatial Distribution of Water resources.pptx
PPTX
Cell cycle ( mitosis and meiosis)Msc.pptx
PPTX
Anaerobic respiration (Fermentation, Types, Pasteur effect, Warburg effect, R...
PPTX
TOOLS OF RECOMBINANT DNA TECHNOLOGY.pptx
PPTX
PHIVOLCS FAULT FINDER grade - 7 power point
PDF
Probiotic bacteria and their importance in aquaculture.pdf
PPTX
VIRULENCE FACTOR OF BACTERIA: STRUCTURAL ELEMENT, ENZYMES, TOXINS...
Chapter 6 Introductory course Biology Camp 1. LECTURE PRESENTATIONS
For CAMPBELL BIOLOGY, NINTH EDITION
Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson
© 2011 Pearson Education, Inc.
Lectures by
Erin Barley
Kathleen Fitzpatrick
A Tour of the Cell
Chapter 6
2. Overview: The Fundamental Units of Life
• All organisms are made of cells
• The cell is the simplest collection of matter
that can be alive
• Cell structure is correlated to cellular function
• All cells are related by their descent from earlier
cells
© 2011 Pearson Education, Inc.
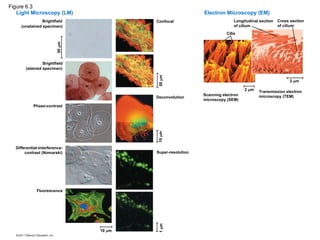
3. Biologists use microscopes and the tools of
biochemistry to study cells
• light microscope (LM): visible light is passed through a specimen and then through glass lenses
– Magnification, the ratio of an object’s image size to its real size; lenses refract the light
– Resolution, the measure of the clarity of the image, or the minimum distance of two
distinguishable points
– Contrast, visible differences in parts of the sample
© 2011 Pearson Education, Inc.
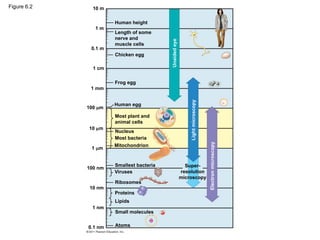
4. Figure 6.2 10 m
1 m
0.1 m
1 cm
1 mm
100 m
10 m
1 m
100 nm
10 nm
1 nm
0.1 nm Atoms
Small molecules
Lipids
Proteins
Ribosomes
Viruses
Smallest bacteria
Mitochondrion
Most bacteria
Nucleus
Most plant and
animal cells
Human egg
Frog egg
Chicken egg
Length of some
nerve and
muscle cells
Human height
Unaided
eye
Light
microscopy
Electron
microscopy
Super-
resolution
microscopy
5. 6. • electron microscopes (EMs): subcellular
structures
– Scanning electron microscopes (SEMs): focus a
beam of electrons onto the surface of a specimen,
providing images that look 3-D
• Transmission electron microscopes (TEMs):
focus a beam of electrons through a specimen
• internal structure of cells
© 2011 Pearson Education, Inc.
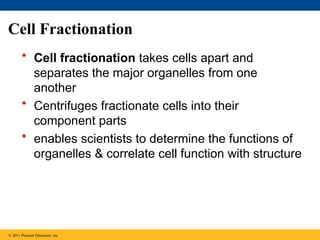
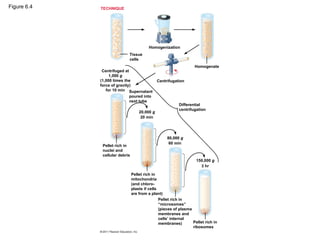
7. Cell Fractionation
• Cell fractionation takes cells apart and
separates the major organelles from one
another
• Centrifuges fractionate cells into their
component parts
• enables scientists to determine the functions of
organelles & correlate cell function with structure
© 2011 Pearson Education, Inc.
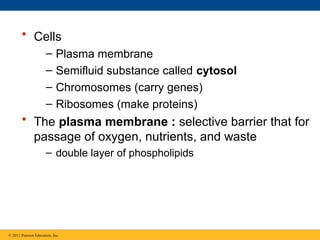
8. 9. • Cells
– Plasma membrane
– Semifluid substance called cytosol
– Chromosomes (carry genes)
– Ribosomes (make proteins)
• The plasma membrane : selective barrier that for
passage of oxygen, nutrients, and waste
– double layer of phospholipids
© 2011 Pearson Education, Inc.
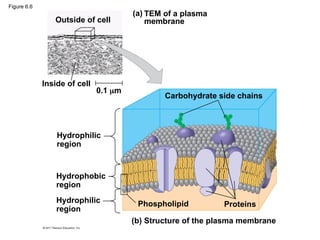
10. Figure 6.6
Outside of cell
Inside of cell
0.1 m
(a) TEM of a plasma
membrane
Hydrophilic
region
Hydrophobic
region
Hydrophilic
region
Carbohydrate side chains
Proteins
Phospholipid
(b) Structure of the plasma membrane
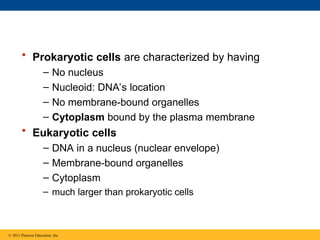
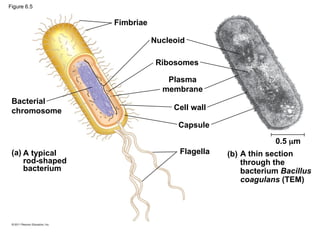
11. • Prokaryotic cells are characterized by having
– No nucleus
– Nucleoid: DNA’s location
– No membrane-bound organelles
– Cytoplasm bound by the plasma membrane
• Eukaryotic cells
– DNA in a nucleus (nuclear envelope)
– Membrane-bound organelles
– Cytoplasm
– much larger than prokaryotic cells
© 2011 Pearson Education, Inc.
12. 13. • Size of Cell: metabolic requirements
– surface area to volume ratio
– surface area increases by n2
, volume increases by n3
• Small cells have a greater surface area relative to volume
© 2011 Pearson Education, Inc.
14. Surface area increases while
total volume remains constant
Total surface area
[sum of the surface areas
(height width) of all box
sides number of boxes]
Total volume
[height width length
number of boxes]
Surface-to-volume
(S-to-V) ratio
[surface area volume]
1
5
6 150 750
1
125
125
1
1.2
6 6
Figure 6.7
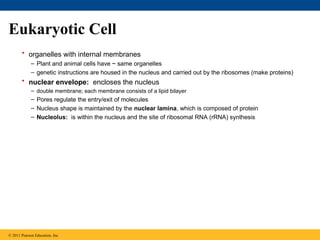
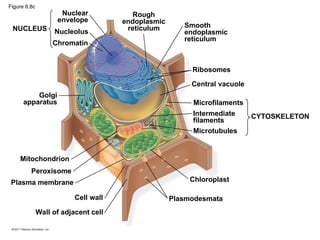
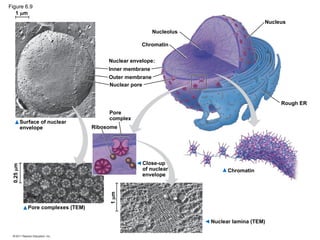
15. Eukaryotic Cell
• organelles with internal membranes
– Plant and animal cells have ~ same organelles
– genetic instructions are housed in the nucleus and carried out by the ribosomes (make proteins)
• nuclear envelope: encloses the nucleus
– double membrane; each membrane consists of a lipid bilayer
– Pores regulate the entry/exit of molecules
– Nucleus shape is maintained by the nuclear lamina, which is composed of protein
– Nucleolus: is within the nucleus and the site of ribosomal RNA (rRNA) synthesis
© 2011 Pearson Education, Inc.
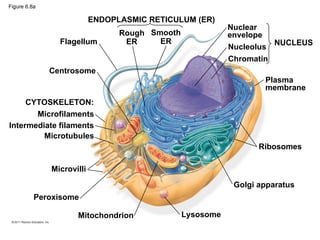
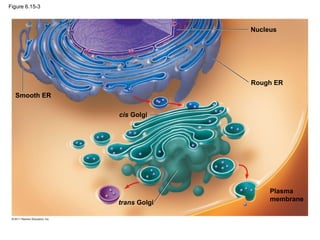
16. Figure 6.8a
ENDOPLASMIC RETICULUM (ER)
Rough
ER
Smooth
ER
Nuclear
envelope
Nucleolus
Chromatin
Plasma
membrane
Ribosomes
Golgi apparatus
Lysosome
Mitochondrion
Peroxisome
Microvilli
Microtubules
Intermediate filaments
Microfilaments
Centrosome
CYTOSKELETON:
Flagellum NUCLEUS
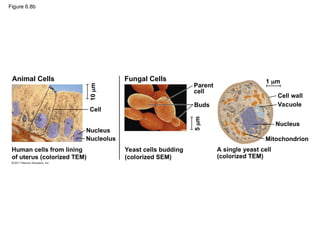
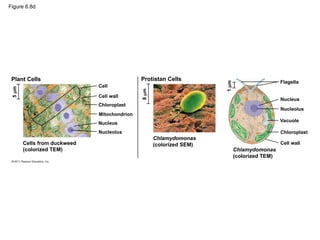
17. 18. 19. Figure 6.8d
Plant Cells
Cells from duckweed
(colorized TEM)
Cell
5
m
Cell wall
Chloroplast
Nucleus
Nucleolus
8
m
Protistan Cells
1
m
Chlamydomonas
(colorized SEM)
Chlamydomonas
(colorized TEM)
Flagella
Nucleus
Nucleolus
Vacuole
Chloroplast
Cell wall
Mitochondrion
20. 21. • Chromosomes: discrete units of DNA
– Chromatin: single DNA molecule associated with
proteins
– chromosomes: condensed chromatin as seen prior to
cell division
• Ribosomes: ribosomal RNA and protein
– protein synthesis in the cytosol (free ribosomes) or
on the outside of the endoplasmic reticulum or
the nuclear envelope (bound ribosomes)
© 2011 Pearson Education, Inc.
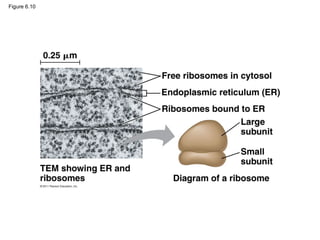
22. Figure 6.10
0.25 m
Free ribosomes in cytosol
Endoplasmic reticulum (ER)
Ribosomes bound to ER
Large
subunit
Small
subunit
Diagram of a ribosome
TEM showing ER and
ribosomes
23. The endomembrane system regulates
protein traffic and performs metabolic
functions in the cell
• Components of the endomembrane system
– Nuclear envelope
– Endoplasmic reticulum
– Golgi apparatus
– Lysosomes
– Vacuoles
– Plasma membrane
• These components are either continuous or
connected via transfer by vesicles
© 2011 Pearson Education, Inc.
24. The Endoplasmic Reticulum: Biosynthetic
Factory
• more than half of the total membrane in
eukaryotic cells
• The ER membrane is continuous with the nuclear
envelope
• There are two distinct regions of ER
– Smooth ER, which lacks ribosomes
– Rough ER, surface is studded with ribosomes
© 2011 Pearson Education, Inc.
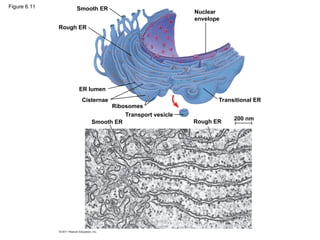
25. Figure 6.11 Smooth ER
Rough ER
ER lumen
Cisternae
Ribosomes
Smooth ER
Transport vesicle
Transitional ER
Rough ER
200 nm
Nuclear
envelope
26. • Smooth ER
– Synthesizes lipids
– Metabolizes carbohydrates
– Detoxifies drugs and poisons
– Stores calcium ions
• Rough ER
– Has bound ribosomes, which secrete glycoproteins (proteins
covalently bonded to carbohydrates)
– Distributes transport vesicles, proteins surrounded by membranes
– Is a membrane factory for the cell
© 2011 Pearson Education, Inc.
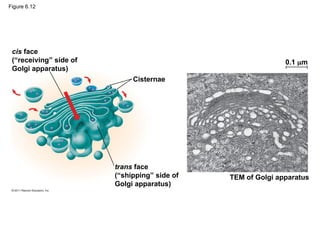
27. • flattened membranous sacs called cisternae
• Functions
– Modifies products of the ER
– Manufactures certain macromolecules
– Sorts and packages materials into transport
vesicles
The Golgi Apparatus: Shipping and
Receiving Center
© 2011 Pearson Education, Inc.
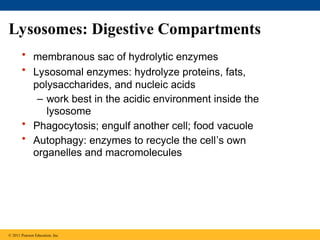
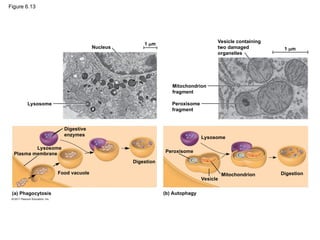
28. 29. Lysosomes: Digestive Compartments
• membranous sac of hydrolytic enzymes
• Lysosomal enzymes: hydrolyze proteins, fats,
polysaccharides, and nucleic acids
– work best in the acidic environment inside the
lysosome
• Phagocytosis; engulf another cell; food vacuole
• Autophagy: enzymes to recycle the cell’s own
organelles and macromolecules
© 2011 Pearson Education, Inc.
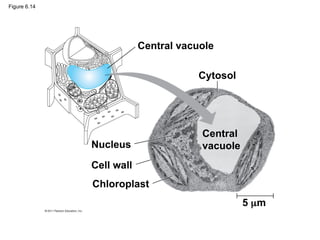
30. 31. Vacuoles: Diverse Maintenance
Compartments
• A plant cell or fungal cell may have one or
several vacuoles, derived from endoplasmic
reticulum and Golgi apparatus
– Food vacuoles: phagocytosis
– Contractile vacuoles: (freshwater protists) pump
excess water out of cells
– Central vacuoles (mature plant cells) hold organic
compounds and water
© 2011 Pearson Education, Inc.
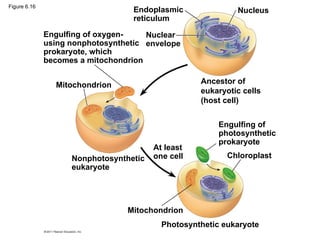
32. 33. 34. Mitochondria and chloroplasts change
energy from one form to another
• Mitochondria: cellular respiration;
• use oxygen ATP
• Chloroplasts (plants, algae) photosynthesis
• Peroxisomes: oxidative organelles
• Mitochondria & chloroplasts: similar to bacteria
– double membrane
– free ribosomes and circular DNA molecules
– Grow and reproduce somewhat independently in cells
© 2011 Pearson Education, Inc.
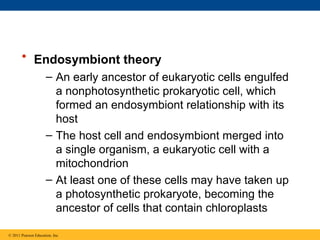
35. • Endosymbiont theory
– An early ancestor of eukaryotic cells engulfed
a nonphotosynthetic prokaryotic cell, which
formed an endosymbiont relationship with its
host
– The host cell and endosymbiont merged into
a single organism, a eukaryotic cell with a
mitochondrion
– At least one of these cells may have taken up
a photosynthetic prokaryote, becoming the
ancestor of cells that contain chloroplasts
© 2011 Pearson Education, Inc.
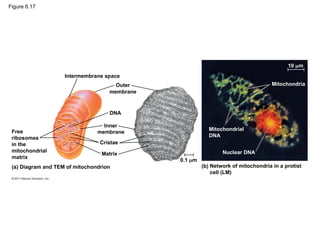
36. 37. Mitochondria: Chemical Energy Conversion
• They have a smooth outer membrane and an
inner membrane folded into cristae
– inner membrane creates two compartments:
intermembrane space and mitochondrial matrix
– Some metabolic steps of cellular respiration are
catalyzed in the mitochondrial matrix
– Cristae present a large surface area for enzymes that
synthesize ATP
© 2011 Pearson Education, Inc.
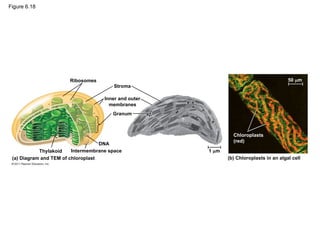
38. 39. Chloroplasts: Capture of Light Energy
• Chloroplasts contain the green pigment chlorophyll, as well as
enzymes and other molecules that function in photosynthesis
• Chloroplasts are found in leaves and other green organs of plants
and in algae
• Chloroplast structure includes
– Thylakoids, membranous sacs, stacked to form a granum
– Stroma, the internal fluid
• The chloroplast is one of a group of plant organelles, called plastids
© 2011 Pearson Education, Inc.
40. Figure 6.18
Ribosomes
Stroma
Inner and outer
membranes
Granum
1 m
Intermembrane space
Thylakoid
(a) Diagram and TEM of chloroplast (b) Chloroplasts in an algal cell
Chloroplasts
(red)
50 m
DNA
41. Peroxisomes: Oxidation
• Peroxisomes are specialized metabolic
compartments bounded by a single membrane
• produce hydrogen peroxide water
• perform reactions with many different functions
• How peroxisomes are related to other organelles
is still unknown
© 2011 Pearson Education, Inc.
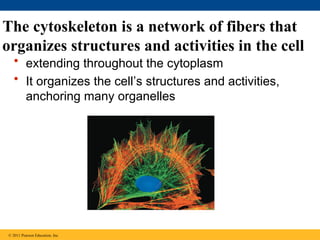
42. The cytoskeleton is a network of fibers that
organizes structures and activities in the cell
• extending throughout the cytoplasm
• It organizes the cell’s structures and activities,
anchoring many organelles
© 2011 Pearson Education, Inc.
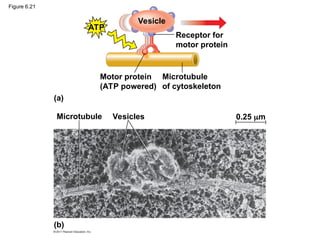
43. Roles of the Cytoskeleton:
Support and Motility
• support the cell and maintain its shape
• It interacts with motor proteins to produce
motility
• Inside the cell, vesicles can travel along
“monorails” provided by the cytoskeleton
• Recent evidence suggests that the cytoskeleton
may help regulate biochemical activities
© 2011 Pearson Education, Inc.
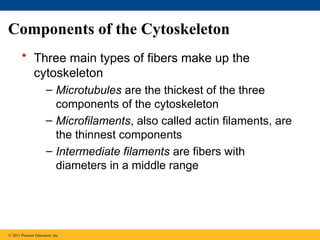
44. 45. Components of the Cytoskeleton
• Three main types of fibers make up the
cytoskeleton
– Microtubules are the thickest of the three
components of the cytoskeleton
– Microfilaments, also called actin filaments, are
the thinnest components
– Intermediate filaments are fibers with
diameters in a middle range
© 2011 Pearson Education, Inc.
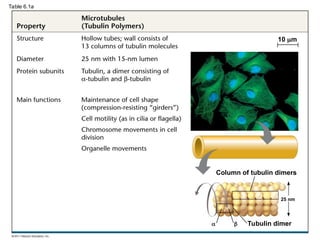
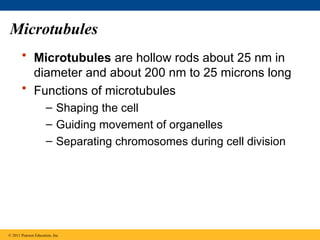
46. 47. 48. 49. Microtubules
• Microtubules are hollow rods about 25 nm in
diameter and about 200 nm to 25 microns long
• Functions of microtubules
– Shaping the cell
– Guiding movement of organelles
– Separating chromosomes during cell division
© 2011 Pearson Education, Inc.
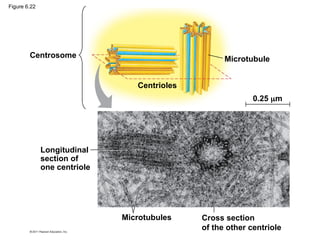
50. Centrosomes and Centrioles
• In many cells, microtubules grow out from a
centrosome near the nucleus
• The centrosome is a “microtubule-organizing
center”
• In animal cells, the centrosome has a pair of
centrioles, each with nine triplets of
microtubules arranged in a ring
© 2011 Pearson Education, Inc.
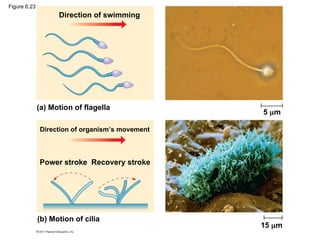
51. 52. Cilia and Flagella
• Microtubules control the beating of cilia and flagella, locomotor appendages
of some cells
• Cilia and flagella differ in their beating patterns
• Cilia and flagella share a common structure
– A core of microtubules sheathed by the plasma membrane
– basal body : anchors the cilium or flagellum
– Dynein: motor protein, which drives the bending movements of a cilium or
flagellum
© 2011 Pearson Education, Inc.
53. Direction of swimming
(b) Motion of cilia
Direction of organism’s movement
Power stroke Recovery stroke
(a) Motion of flagella
5 m
15 m
Figure 6.23
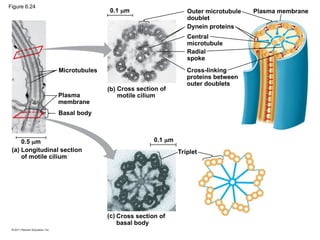
54. Microtubules
Plasma
membrane
Basal body
Longitudinal section
of motile cilium
(a)
0.5 m 0.1 m
0.1 m
(b) Cross section of
motile cilium
Outer microtubule
doublet
Dynein proteins
Central
microtubule
Radial
spoke
Cross-linking
proteins between
outer doublets
Plasma membrane
Triplet
(c) Cross section of
basal body
Figure 6.24
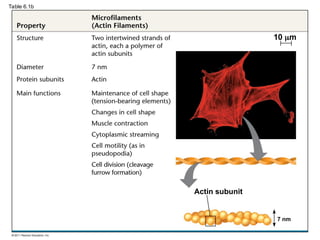
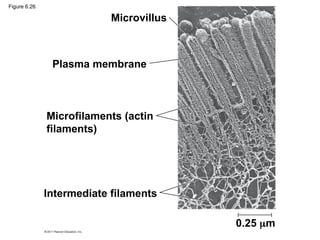
55. Microfilaments (Actin Filaments)
• solid rods about 7 nm in diameter, built as a
twisted double chain of actin subunits
– bear tension, resisting pulling forces within the cell
– Cortex: 3-D network for shape
– make up the core of microvilli of intestinal cells
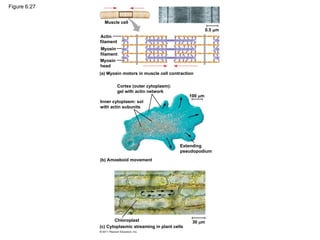
• Myosin: in microfilaments for cellular motility
– In muscle cells, actin filaments are parallel
– Thicker filaments (myosin) with the thinner actin fibers
© 2011 Pearson Education, Inc.
56. 57. 58. • Localized contraction brought about by actin and myosin also drives
amoeboid movement
• Pseudopodia (cellular extensions) extend and contract through the
reversible assembly and contraction of actin subunits into
microfilaments
• Cytoplasmic streaming is a circular flow of cytoplasm within cells
– speeds distribution of materials within the cell
• In plant cells, actin-myosin interactions and sol-gel transformations
drive cytoplasmic streaming
© 2011 Pearson Education, Inc.
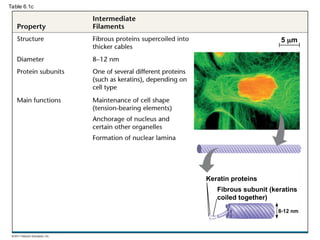
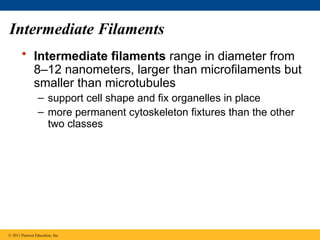
59. Intermediate Filaments
• Intermediate filaments range in diameter from
8–12 nanometers, larger than microfilaments but
smaller than microtubules
– support cell shape and fix organelles in place
– more permanent cytoskeleton fixtures than the other
two classes
© 2011 Pearson Education, Inc.
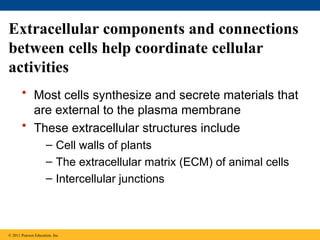
60. Extracellular components and connections
between cells help coordinate cellular
activities
• Most cells synthesize and secrete materials that
are external to the plasma membrane
• These extracellular structures include
– Cell walls of plants
– The extracellular matrix (ECM) of animal cells
– Intercellular junctions
© 2011 Pearson Education, Inc.
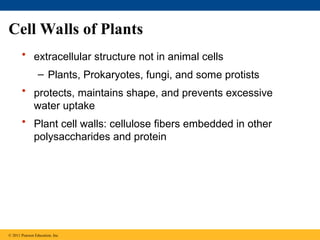
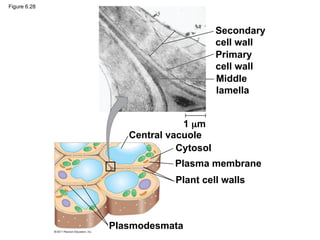
61. Cell Walls of Plants
• extracellular structure not in animal cells
– Plants, Prokaryotes, fungi, and some protists
• protects, maintains shape, and prevents excessive
water uptake
• Plant cell walls: cellulose fibers embedded in other
polysaccharides and protein
© 2011 Pearson Education, Inc.
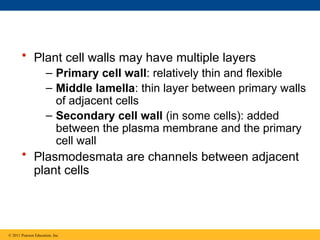
62. • Plant cell walls may have multiple layers
– Primary cell wall: relatively thin and flexible
– Middle lamella: thin layer between primary walls
of adjacent cells
– Secondary cell wall (in some cells): added
between the plasma membrane and the primary
cell wall
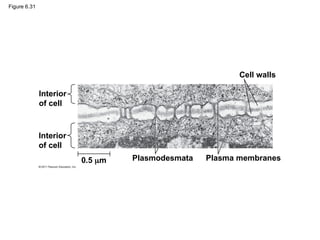
• Plasmodesmata are channels between adjacent
plant cells
© 2011 Pearson Education, Inc.
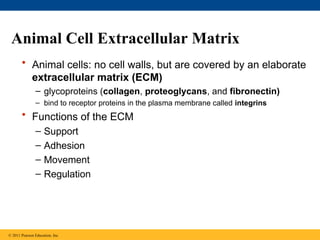
63. 64. 65. Animal Cell Extracellular Matrix
• Animal cells: no cell walls, but are covered by an elaborate
extracellular matrix (ECM)
– glycoproteins (collagen, proteoglycans, and fibronectin)
– bind to receptor proteins in the plasma membrane called integrins
• Functions of the ECM
– Support
– Adhesion
– Movement
– Regulation
© 2011 Pearson Education, Inc.
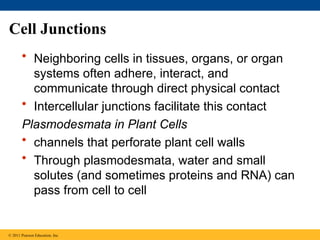
66. 67. Cell Junctions
• Neighboring cells in tissues, organs, or organ
systems often adhere, interact, and
communicate through direct physical contact
• Intercellular junctions facilitate this contact
Plasmodesmata in Plant Cells
• channels that perforate plant cell walls
• Through plasmodesmata, water and small
solutes (and sometimes proteins and RNA) can
pass from cell to cell
© 2011 Pearson Education, Inc.
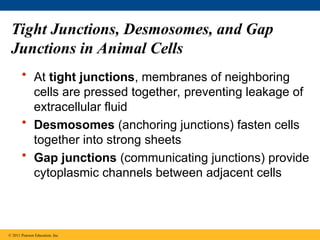
68. 69. Tight Junctions, Desmosomes, and Gap
Junctions in Animal Cells
• At tight junctions, membranes of neighboring
cells are pressed together, preventing leakage of
extracellular fluid
• Desmosomes (anchoring junctions) fasten cells
together into strong sheets
• Gap junctions (communicating junctions) provide
cytoplasmic channels between adjacent cells
© 2011 Pearson Education, Inc.
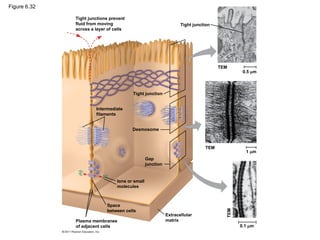
70. Figure 6.32
Tight junctions prevent
fluid from moving
across a layer of cells
Tight junction
Tight junction
TEM
0.5 m
TEM
1 m
TEM
0.1 m
Extracellular
matrix
Plasma membranes
of adjacent cells
Space
between cells
Ions or small
molecules
Desmosome
Intermediate
filaments
Gap
junction
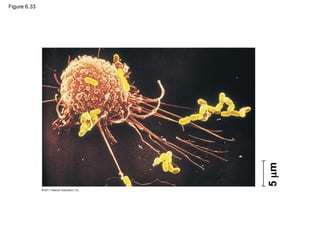
71. The Cell: A Living Unit Greater Than the
Sum of Its Parts
• Cells rely on the integration of structures and
organelles in order to function
• For example, a macrophage’s ability to destroy
bacteria involves the whole cell, coordinating
components such as the cytoskeleton,
lysosomes, and plasma membrane
© 2011 Pearson Education, Inc.
72. 73. Editor's Notes #2 For the Discovery Video Cells, go to Animation and Video Files.
#4 Figure 6.2 The size range of cells. #5 Figure 6.3 Exploring: Microscopy #8 Figure 6.4 Research Method: Cell Fractionation #10 Figure 6.6 The plasma membrane. #12 Figure 6.5 A prokaryotic cell. #14 Figure 6.7 Geometric relationships between surface area and volume. #16 Figure 6.8 Exploring: Eukaryotic Cells #17 Figure 6.8 Exploring: Eukaryotic Cells #18 Figure 6.8 Exploring: Eukaryotic Cells #19 Figure 6.8 Exploring: Eukaryotic Cells #20 Figure 6.9 The nucleus and its envelope. #22 Figure 6.10 Ribosomes. #24 For the Cell Biology Video ER and Mitochondria in Leaf Cells, go to Animation and Video Files.
#25 Figure 6.11 Endoplasmic reticulum (ER). #27 For the Cell Biology Video ER to Golgi Traffic, go to Animation and Video Files.
For the Cell Biology Video Golgi Complex in 3D, go to Animation and Video Files.
For the Cell Biology Video Secretion From the Golgi, go to Animation and Video Files.
#28 Figure 6.12 The Golgi apparatus. #30 Figure 6.13 Lysosomes. #32 Figure 6.14 The plant cell vacuole. #33 Figure 6.15 Review: relationships among organelles of the endomembrane system. #34 For the Cell Biology Video ER and Mitochondria in Leaf Cells, go to Animation and Video Files.
For the Cell Biology Video Mitochondria in 3D, go to Animation and Video Files.
For the Cell Biology Video Chloroplast Movement, go to Animation and Video Files.
#36 Figure 6.16 The endosymbiont theory of the origin of mitochondria and chloroplasts in eukaryotic cells. #38 Figure 6.17 The mitochondrion, site of cellular respiration. #40 Figure 6.18 The chloroplast, site of photosynthesis. #42 For the Cell Biology Video The Cytoskeleton in a Neuron Growth Cone, go to Animation and Video Files
For the Cell Biology Video Cytoskeletal Protein Dynamics, go to Animation and Video Files.
#44 Figure 6.21 Motor proteins and the cytoskeleton. #45 For the Cell Biology Video Actin Network in Crawling Cells, go to Animation and Video Files.
For the Cell Biology Video Actin Visualization in Dendrites, go to Animation and Video Files.
#46 Table 6.1 The Structure and Function of the Cytoskeleton #47 Table 6.1 The Structure and Function of the Cytoskeleton #48 Table 6.1 The Structure and Function of the Cytoskeleton #49 For the Cell Biology Video Transport Along Microtubules, go to Animation and Video Files.
For the Cell Biology Video Movement of Organelles in Vivo, go to Animation and Video Files.
For the Cell Biology Video Movement of Organelles in Vitro, go to Animation and Video Files.
#51 Figure 6.22 Centrosome containing a pair of centrioles. #53 Figure 6.23 A comparison of the beating of flagella and motile cilia. #54 Figure 6.24 Structure of a flagellum or motile cilium. #56 Figure 6.26 A structural role of microfilaments. #57 Figure 6.27 Microfilaments and motility. #59 For the Cell Biology Video Interphase Microtubule Dynamics, go to Animation and Video Files.
For the Cell Biology Video Microtubule Sliding in Flagellum Movement, go to Animation and Video Files.
For the Cell Biology Video Microtubule Dynamics, go to Animation and Video Files.
#60 For the Cell Biology Video Ciliary Motion, go to Animation and Video Files.
#62 For the Cell Biology Video E-cadherin Expression, go to Animation and Video Files.
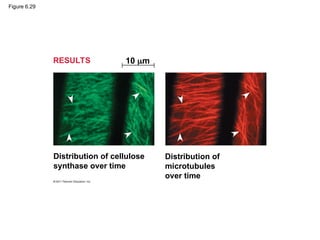
#63 Figure 6.28 Plant cell walls. #64 Figure 6.29 Inquiry: What role do microtubules play in orienting deposition of cellulose in cell walls? #65 For the Cell Biology Video Cartoon Model of a Collagen Triple Helix, go to Animation and Video Files.
For the Cell Biology Video Staining of the Extracellular Matrix, go to Animation and Video Files.
For the Cell Biology Video Fibronectin Fibrils, go to Animation and Video Files.
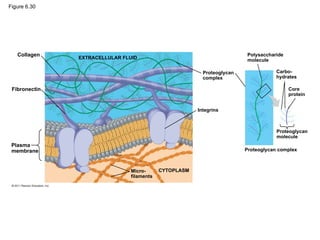
#66 Figure 6.30 Extracellular matrix (ECM) of an animal cell. #68 Figure 6.31 Plasmodesmata between plant cells. #70 Figure 6.32 Exploring: Cell Junctions in Animal Tissues #72 Figure 6.33 The emergence of cellular functions. #73 Figure 6.UN01 Summary table, Concepts 6.3–6.5




![Surface area increases while
total volume remains constant
Total surface area
[sum of the surface areas
(height width) of all box
sides number of boxes]
Total volume
[height width length
number of boxes]
Surface-to-volume
(S-to-V) ratio
[surface area volume]
1
5
6 150 750
1
125
125
1
1.2
6 6
Figure 6.7](https://image.slidesharecdn.com/06-cells-250825072452-c139ffa4/85/Chapter-6-Introductory-course-Biology-Camp-14-320.jpg)