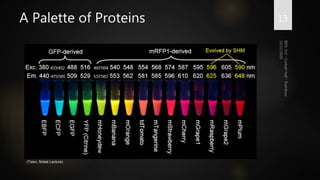
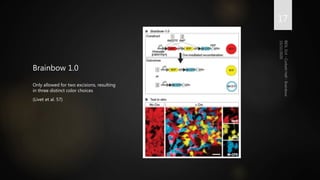
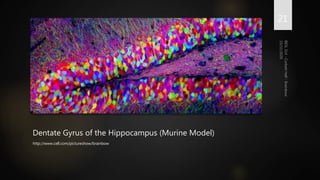
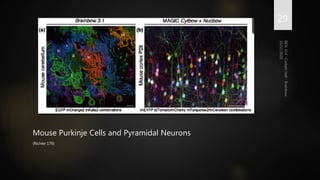
Brainbow is a technique that uses fluorescent proteins to map neural connections in the brain. It involves genetically engineering mice so that individual neurons express different combinations of colors. This allows their connections to be traced visually. The technique was improved over time by incorporating additional genetic tools. Brainbow has been applied to map connectomes in mice, fish, flies and more. It provides a way to visualize neural circuits and how they change over time or with conditions like disease.