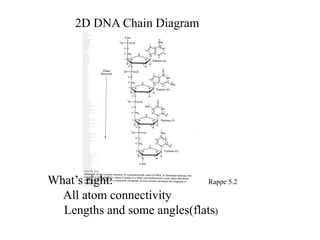
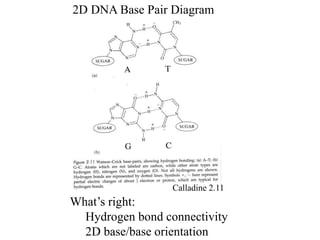
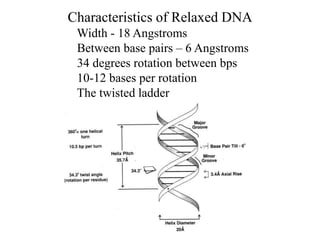
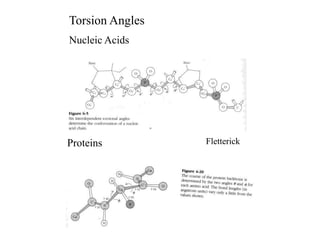
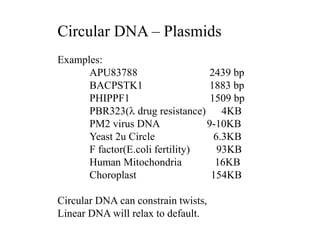
1. The document outlines topics related to DNA geometry including one-dimensional (1D), two-dimensional (2D), and three-dimensional (3D) representations as well as circular DNA topology and measures like writhe, twist, and linking number.
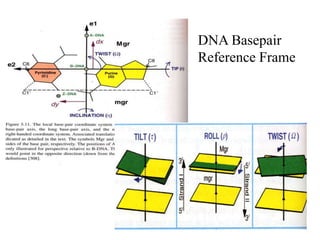
2. It describes the NAB software package for modeling DNA and its energy model that considers torsion, curvature, twist, tilt, and roll.
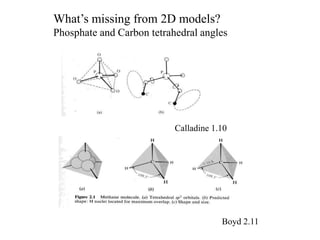
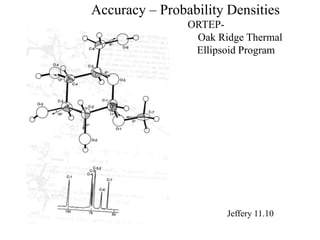
3. The document discusses accuracy issues in modeling DNA and visualization techniques like overlapped and narrowed views as well as probability densities from methods like NMR.






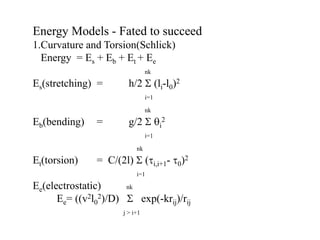
![. Twist, Tilt and Roll
E= .5kBT [ k1 [k0(Twi-Tw0) + (Roi-Ro0)]2
+k2 [k0(Twi-Tw0) - (Roi-Ro0)]2
+k3 [ (Tli-Tl0) ]2 ]](https://image.slidesharecdn.com/beard1-231026181450-313e6054/85/beard1-ppt-17-320.jpg)