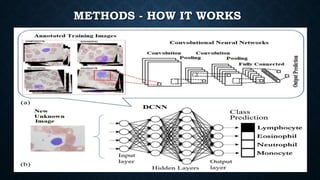
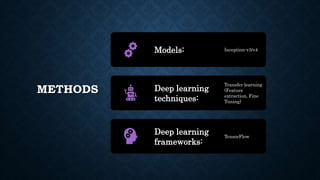
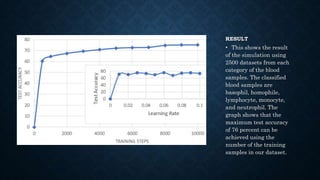
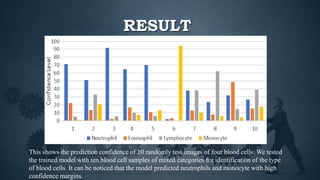
This document summarizes research on using deep convolutional neural networks to automatically analyze microscopy images. The goals are to expedite the analysis of high-content microscopy data and automate tasks like cell counting and classification. The researchers trained and tested models using TensorFlow on microscopy images to classify cells, achieving over 75% accuracy. This level of automation could benefit biological research by reducing human errors and speeding up analysis of large image datasets.




![THE BUILDING
BLOCKS OF DCNN
• Multi-Layer Perceptrons
(MLPs) are among the most
fundamental building blocks
in Artificial Neural Networks
(ANNs). It refers to a set of
computational models that
are loosely inspired by the
human brain. In general,
they consist of two important
elements, namely, artificial
neurons (nodes) and
synapses (weights) that
connect them. LeCun, [18]](https://image.slidesharecdn.com/yaserbanadaki-spiepresentationslide-210208215626/85/Automated-Analysis-of-Microscopy-Images-using-Deep-Convolutional-Neural-Network-8-320.jpg)