The document outlines a group assignment focused on enzyme structure, classification, and mechanisms of action, including preparation guidelines for a PowerPoint presentation. Key learning objectives include defining enzymes, explaining their functions, and understanding their specificity and classification. Additionally, it covers various factors affecting enzyme activity, including cofactors, environmental conditions, and inhibitors.

![Aspect Component Score
Group ID and leader
A B
C
D E
Subject
matter
Introduction 5
Measuring [enzyme] 5
Assay methods 15
Important additional
details
5
Conclusion &
references
5
Presentation
quality
Slide quality &
content
5
Slide numbering &
editorials
5
Presenter
composure, voice
projection & clarity
5
Presenter
appearance
5
Time keeping 5
Total 60
3](https://image.slidesharecdn.com/as116enzymes1-240818165944-04e8bb2e/75/AS116-Enzymes-mode-of-action-of-enzymes-1-ppt-3-2048.jpg)

![Importance
• Enzymes play an important role in Metabolism,
Diagnosis & Therapeutics
• All biochemical reactions are catalyzed by enzymes
• [Enzyme] in blood is of diagnostic significance e.g
good indicator of disease e.g myocardial infarction
• Enzymes can be used therapeutically
6](https://image.slidesharecdn.com/as116enzymes1-240818165944-04e8bb2e/75/AS116-Enzymes-mode-of-action-of-enzymes-1-ppt-6-2048.jpg)




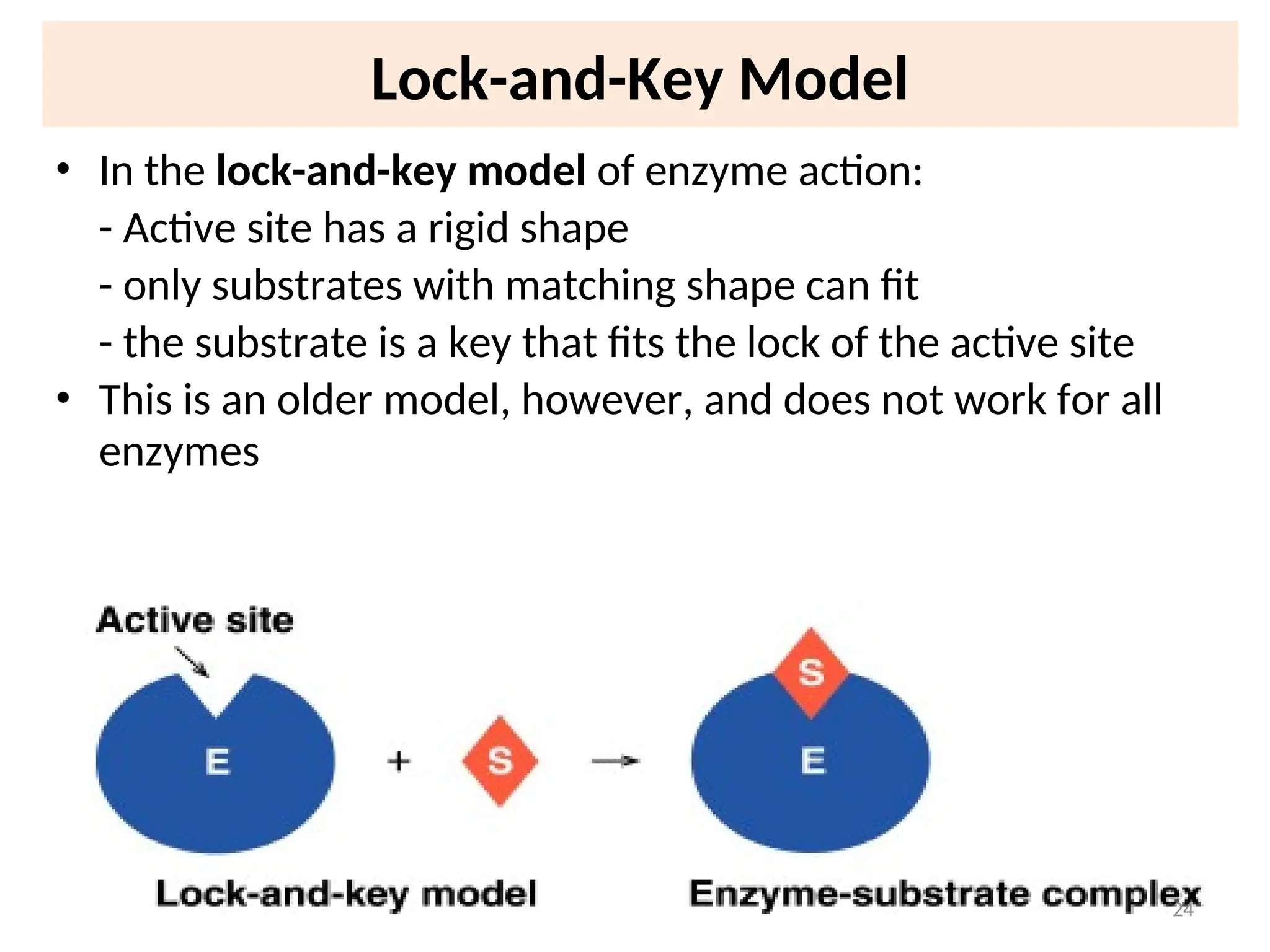
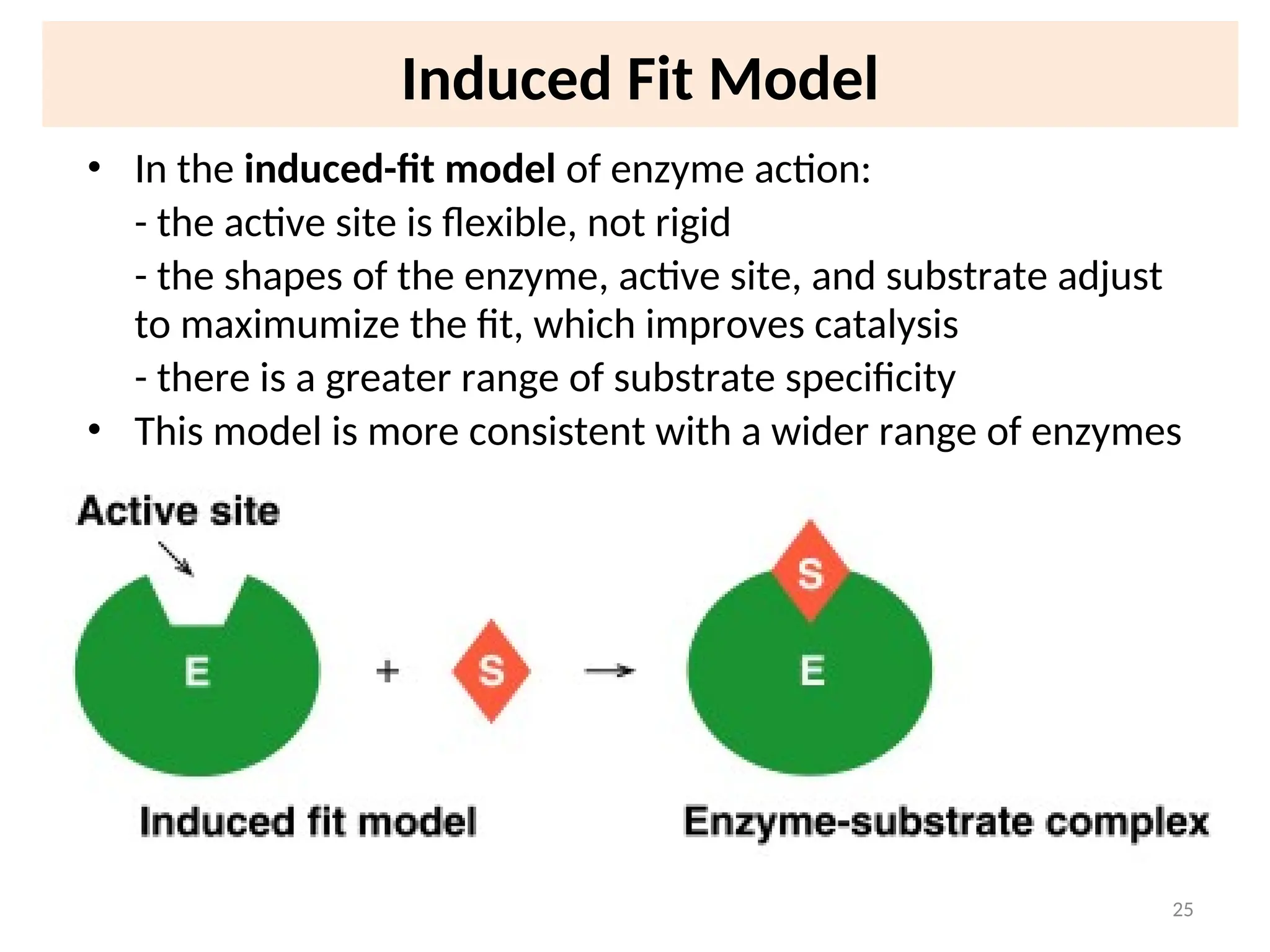
![31
Environmental Conditions
Environmental Conditions
1. Extreme temperatures;
– the most dangerous
– high temps may denature (unfold) the enzyme
2. pH (most like 6 - 8 pH near neutral)
3. Substrate concentration [S]
31](https://image.slidesharecdn.com/as116enzymes1-240818165944-04e8bb2e/75/AS116-Enzymes-mode-of-action-of-enzymes-1-ppt-27-2048.jpg)




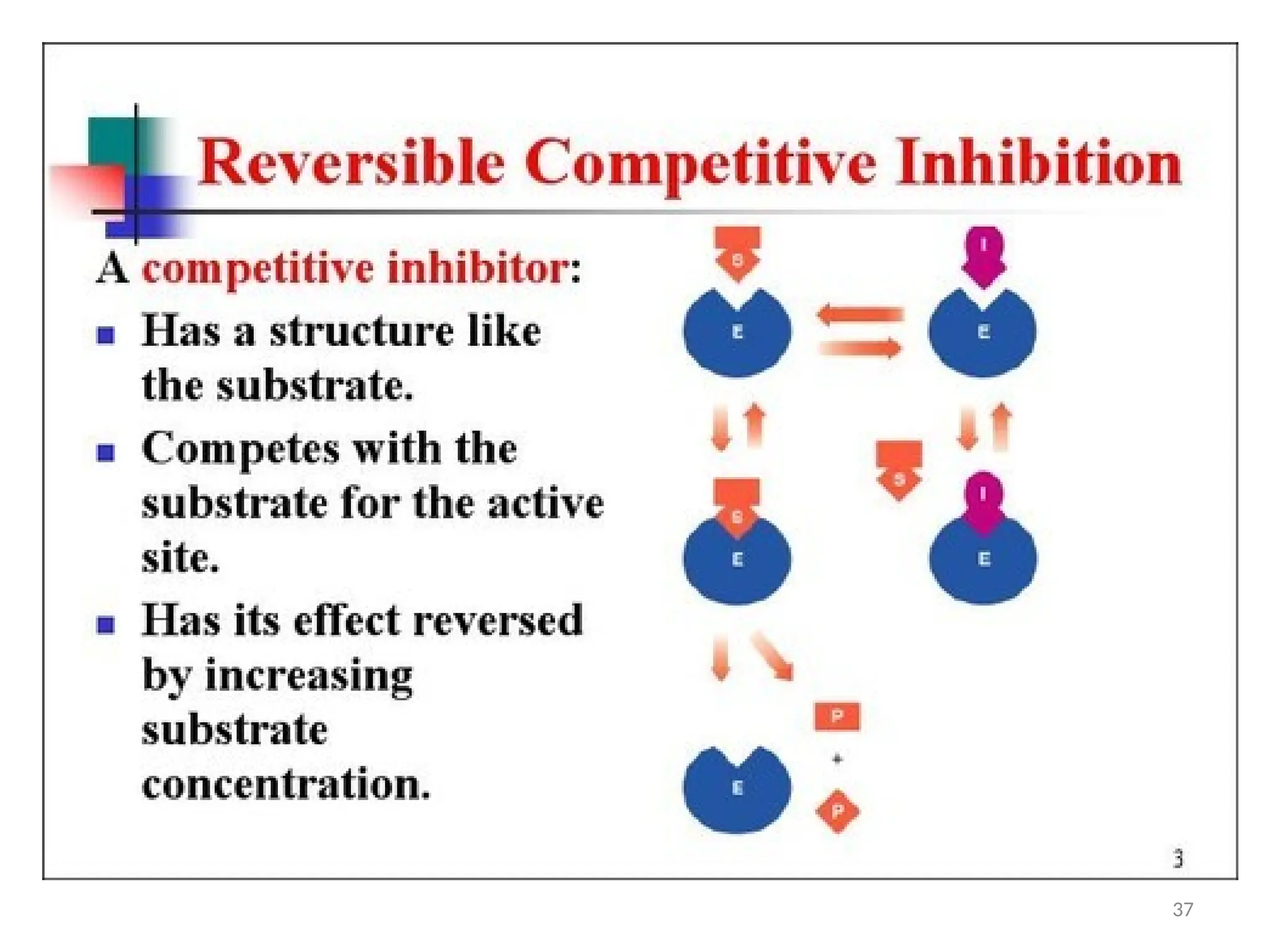
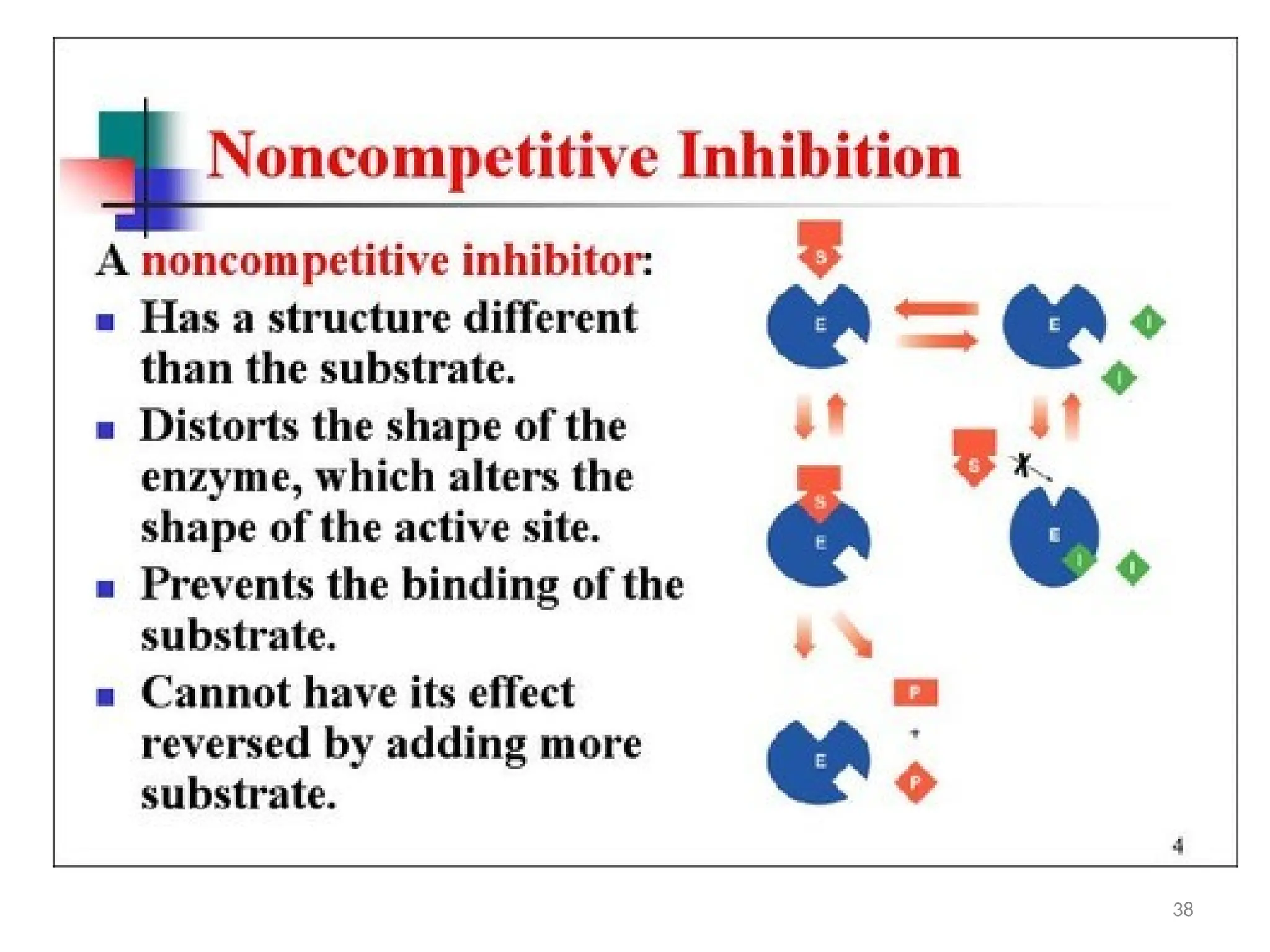
![Irreversible inhibition:
- Suicide inactivators;
•These molecules bind permanently with the
enzyme;
•Reduce the [enzyme]
•Example, cyanide irreversibly inhibits the
enzyme cytochrome oxidase (respiration)
•If this cannot be used, death will occur
39](https://image.slidesharecdn.com/as116enzymes1-240818165944-04e8bb2e/75/AS116-Enzymes-mode-of-action-of-enzymes-1-ppt-34-2048.jpg)