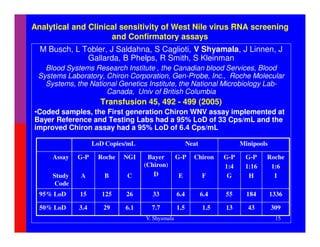
Venkatakrishna Shyamala has extensive experience in molecular diagnostics and research and development. She has held leadership roles at several companies developing molecular diagnostic tests and was involved in the development, validation, and implementation of alternative nucleic acid tests for HCV and WNV screening at Chiron Corporation. She has also consulted for Novartis Diagnostics and contributed to establishing standards for molecular diagnostic methods through her work with CLSI.