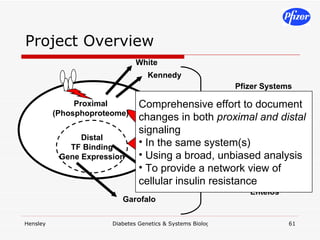
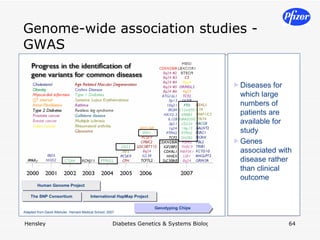
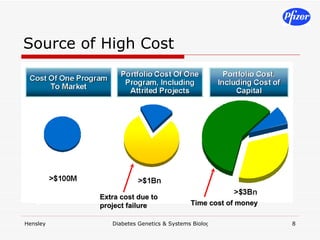
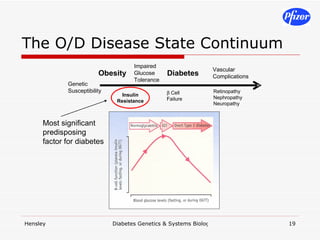
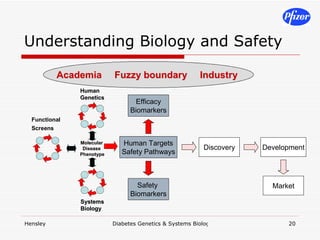
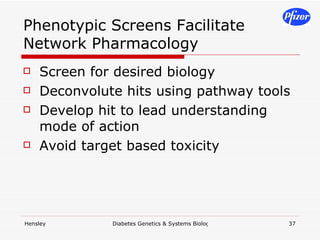
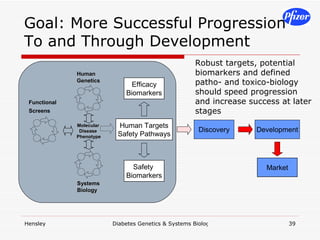
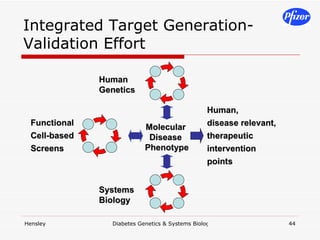
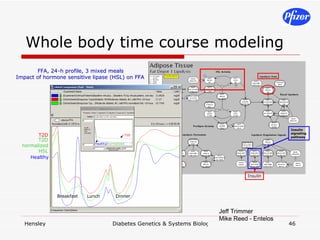
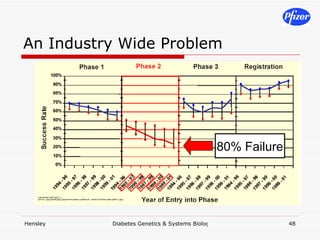
The document discusses using systems biology and genetics approaches to improve drug discovery for diabetes. It argues that current approaches have high failure rates due to a lack of understanding disease biology. It proposes using multi-disciplinary experiments and computational modeling to map signaling networks, identify modulation points, and predict drug targets. This would require new academic-industry partnerships and integrating various omics data to better understand disease mechanisms and improve drug development outcomes.























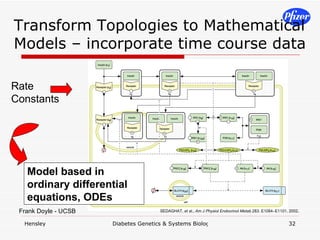
![Pre-existing Knowledge Time course data phosphorylation tf binding expression profiling Pathway knowledge Other knowledge interactome, etc. metabolome 0.0 0.2 0.4 0.6 0.8 1.0 1.2 0 10 20 30 40 50 Time [min] Amount normalized with 5 min . 0.0 0.2 0.4 0.6 0.8 1.0 1.2 0 10 20 30 40 50 Time [min] Amount normalized with 5 min .](https://image.slidesharecdn.com/DiabetesSystemsBiologyAndGeneticsV6-122775687268-phpapp03/85/Diabetes-Systems-Biology-And-Genetics-V6-28-320.jpg)




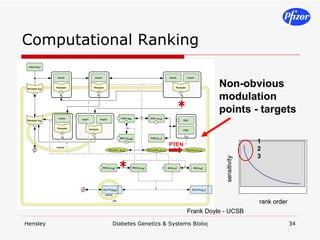














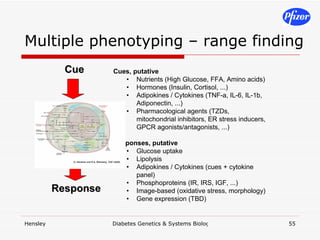



![Targeted CSR stimuli*time points*measurements*inhibitors*[I]*cell types (n=replicates)](https://image.slidesharecdn.com/DiabetesSystemsBiologyAndGeneticsV6-122775687268-phpapp03/85/Diabetes-Systems-Biology-And-Genetics-V6-59-320.jpg)