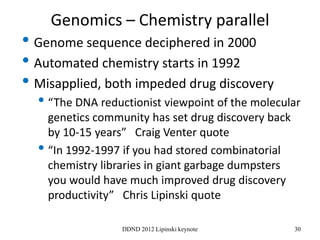
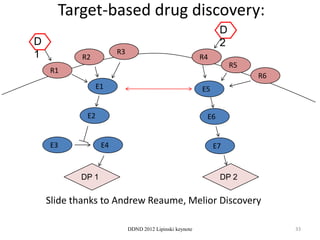
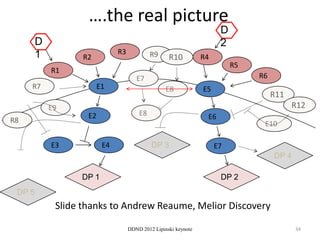
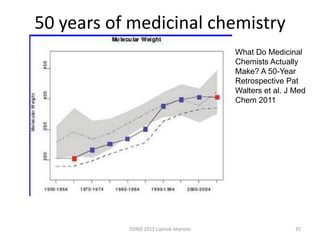
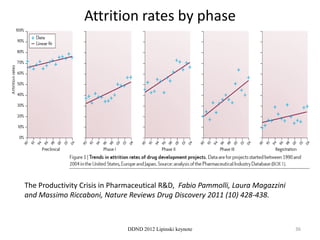
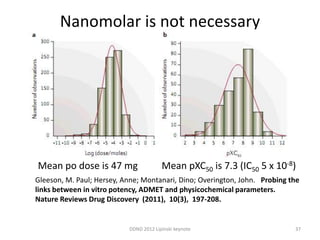
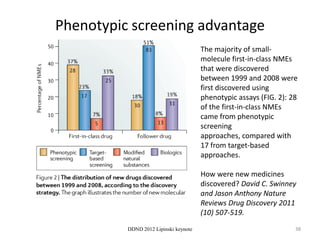
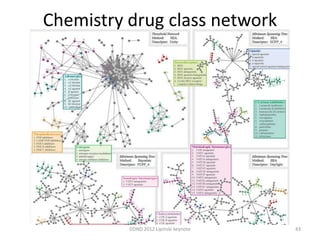
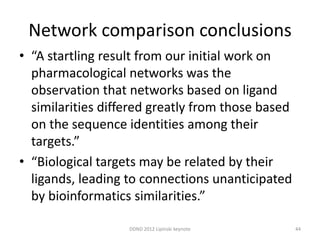
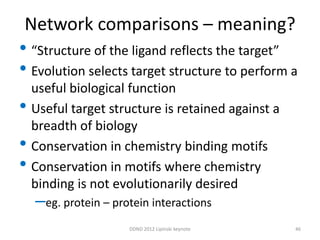
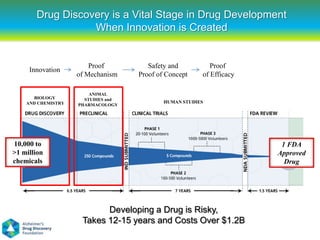
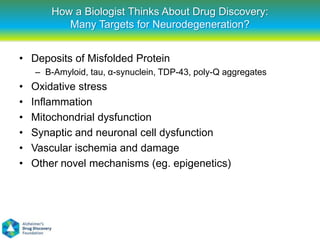
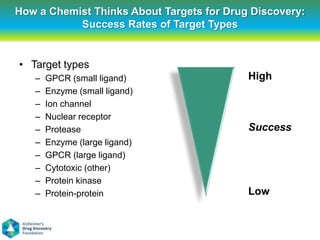
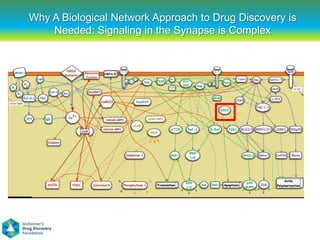
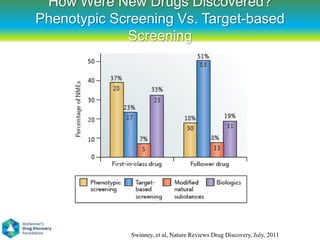
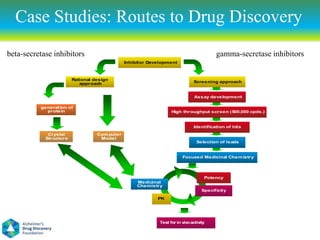
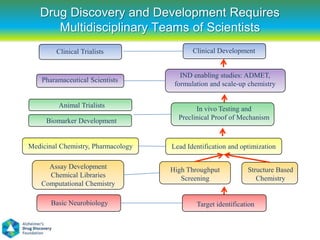
This document summarizes a keynote presentation on the state and future of drug discovery. The presentation discusses: 1) The "valley of death" between academic research and clinical development due to a lack of translational skills and partially flawed academic targets; 2) How genomics and high-throughput screening have set back drug discovery by prioritizing a reductionist approach; and 3) How phenotypic screening and considering biological networks may improve success rates over the traditional target-based approach. The keynote argues for improved target validation and considering the relationships between biological and chemical networks.







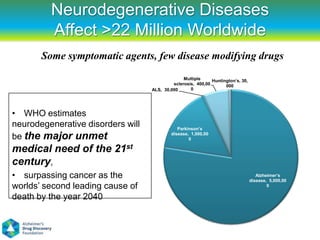








![Translational valley of death
"curing disease is a byproduct of the [NIH] system and not a goal," says
FasterCures' Simon. Most scientists don't want to and don't have the skills to
translate a discovery into a treatment; researchers at a dedicated center would
try to do that full-time.
DDND 2012 Lipinski keynote 24](https://image.slidesharecdn.com/sundayfillet-lipinski-120223151601-phpapp02/85/Sunday-fillet-lipinski-24-320.jpg)