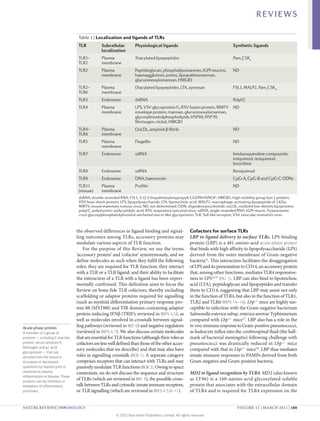
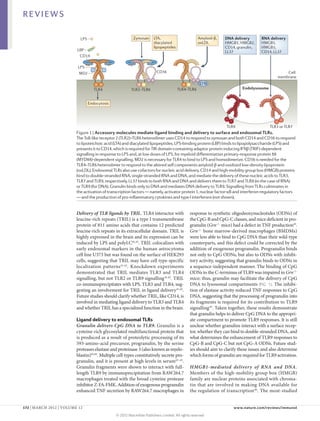
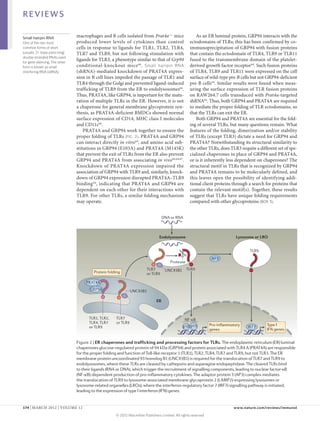
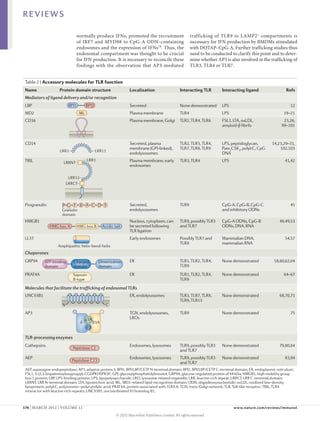
1) Accessory proteins are required for the proper biosynthesis, activation, and function of Toll-like receptors (TLRs). These proteins can act as ligand-recognition and delivery cofactors, chaperones, or proteins involved in TLR trafficking.
2) Key accessory proteins for surface TLRs include LBP, which facilitates LPS recognition by TLR4, and MD2, which associates with TLR4 and is essential for its response to LPS. CD36 also mediates TLR4 and TLR6 responses to oxidized LDL.
3) Accessory proteins play important roles in modulating TLR activation and may be useful targets for manipulating TLR pathways for therapeutic applications.