This document discusses the R debugger. It provides examples of using the browser(), debug(), and debugonce() functions to debug R code. The browser allows stepping through code line-by-line and examining variable values. debug() and debugonce() activate debugging for a function. Other debugging topics covered include traceback(), browserText(), and examining the coefficients of linear models.







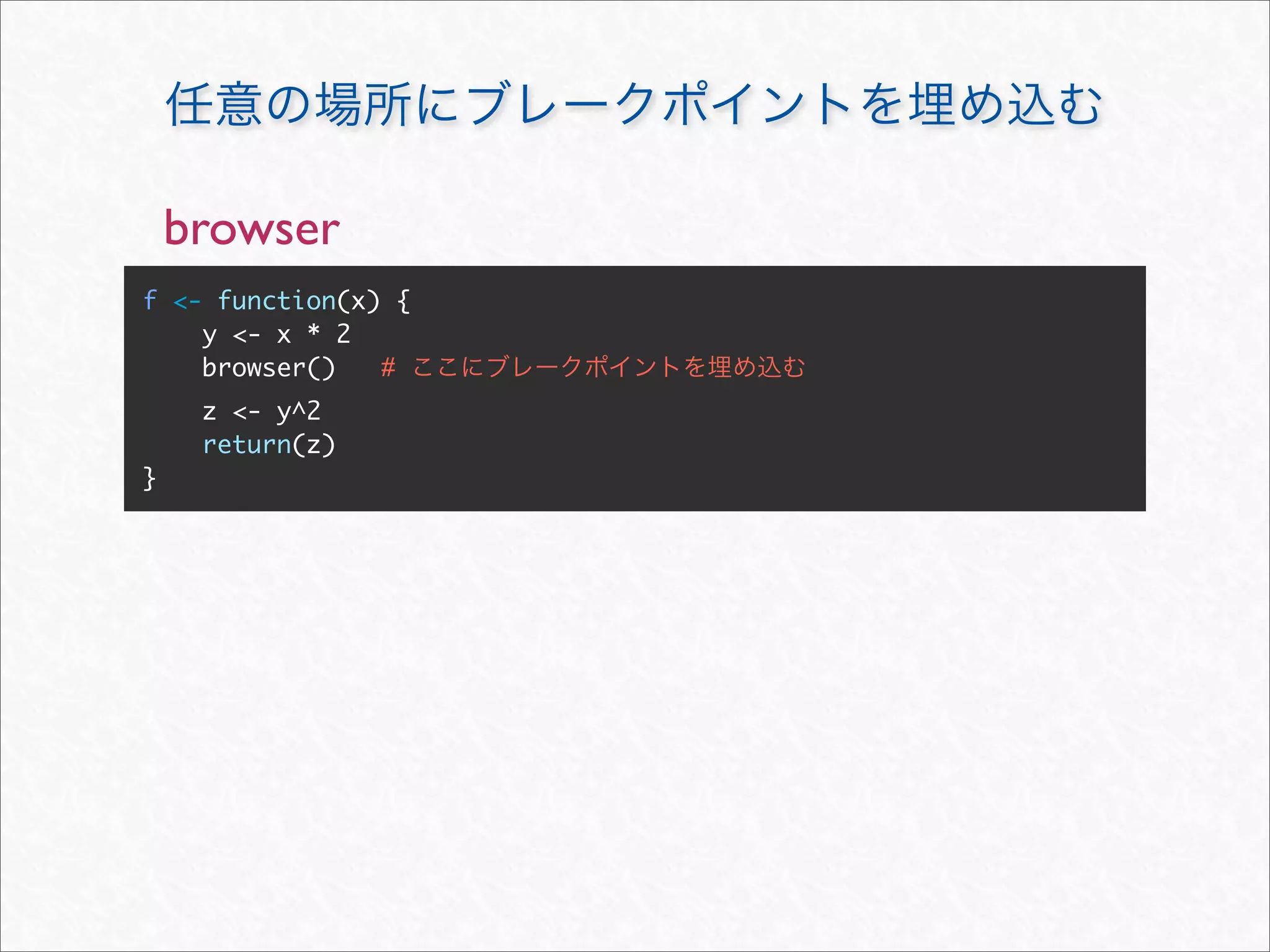
![browser
f <- function(x) { > f(10)
y <- x * 2 Called from: f(10)
Browse[1]>
z <- y^2
return(z)
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-10-2048.jpg)
![browser
f <- function(x) { > f(10)
y <- x * 2 Called from: f(10)
Browse[1]> ls()
z <- y^2 [1] "x" "y"
return(z) Browse[1]>
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-11-2048.jpg)
![browser
f <- function(x) { > f(10)
y <- x * 2 Called from: f(10)
Browse[1]> ls()
z <- y^2 [1] "x" "y"
return(z) Browse[1]> n #
} debug: z <- y^2
Browse[2]>](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-12-2048.jpg)
![browser
f <- function(x) { > f(10)
y <- x * 2 Called from: f(10)
Browse[1]> ls()
z <- y^2 [1] "x" "y"
return(z) Browse[1]> n #
} debug: z <- y^2
Browse[2]> z
Error: object 'z' not found
Browse[2]>
z](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-13-2048.jpg)
![browser
f <- function(x) { > f(10)
y <- x * 2 Called from: f(10)
Browse[1]> ls()
z <- y^2 [1] "x" "y"
return(z) Browse[1]> n #
} debug: z <- y^2
Browse[2]> z
Error: object 'z' not found
Browse[2]> n
debug: return(z)
Browse[2]> z
[1] 400
Browse[2]>
z y^2](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-14-2048.jpg)
![browser
f <- function(x) { > f(10)
y <- x * 2 Called from: f(10)
Browse[1]> ls()
z <- y^2 [1] "x" "y"
return(z) Browse[1]> n #
} debug: z <- y^2
Browse[2]> z
Error: object 'z' not found
Browse[2]> n
debug: return(z)
Browse[2]> z
[1] 400
Browse[2]> z <- 1 # z
Browse[2]> n
[1] 1
>
f(10) z](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-15-2048.jpg)
![debug
f <- function(x) { > debug(f)
y <- x * 2 > f(10)
z <- y^2 debugging in: f(10)
return(z) debug: {
} y <- x * 2
z <- y^2
return(z)
}
Browse[2]> n
debug: y <- x * 2
browser Browse[2]> n
debug: z <- y^2
Browse[2]> n
debug: return(z)
Browse[2]> n
exiting from: f(10)
[1] 400
>](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-16-2048.jpg)
![debugonce
> debug(f) > debugonce(f)
> f(10) > f(10)
debugging in: f(10) debugging in: f(10)
debug: { debug: {
y <- x * 2 y <- x * 2
z <- y^2 z <- y^2
return(z) return(z)
} }
Browse[2]> Q # Browse[2]> Q #
> isdebugged(f) > isdebugged(f)
[1] TRUE [1] FALSE
> f(10) > f(10)
debugging in: f(10) [1] 400
... >
※ undebug(f)](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-17-2048.jpg)
![traceback
f <- function(x) { > f(1:5)
x <- as.character(x) Error in sum(x) : invalid
return(g(x)) 'type' (character) of argument
} > traceback()
3: h(x) h
g <- function(x) { 2: g(x)
if (length(x) > 1) { 1: f(1:10)
return(h(x)) > debug(h)
} else { > f(1:10)
return(x) debugging in: h(x)
} debug: {
} return(sum(x))
}
h <- function(x) { Browse[2]> x
return(sum(x)) [1] "1" "2" "3" "4" "5"
} Browse[2]> sum(x)
Error in sum(x) : invalid
'type' (character) of argument](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-18-2048.jpg)
![browser
f <- function(x) { > f(5) # y = 10
y <- x * 2 [1] 100
browser(expr = y > 10) > f(10) # y = 20
z <- y^2 Called from: f(10)
return(z) Browse[1]>
}
f <- function(x) {
y <- x * 2
if (y > 10) {
browser()
}
z <- y^2
return(z)
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-19-2048.jpg)
![browserText
f <- function(x) { > f(10)
browser("1st bt") Called from: f(10)
Browse[1]> where #
# < >
where 1: f(10)
browser("2nd bt")
}
Browse[1]> browserText()
[1] "1st bt"
Browse[1]> c
Called from: f(10)
Browse[1]> where #
where 1: f(10)
Browse[1]> browserText()
[1] "2nd bt"](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-20-2048.jpg)


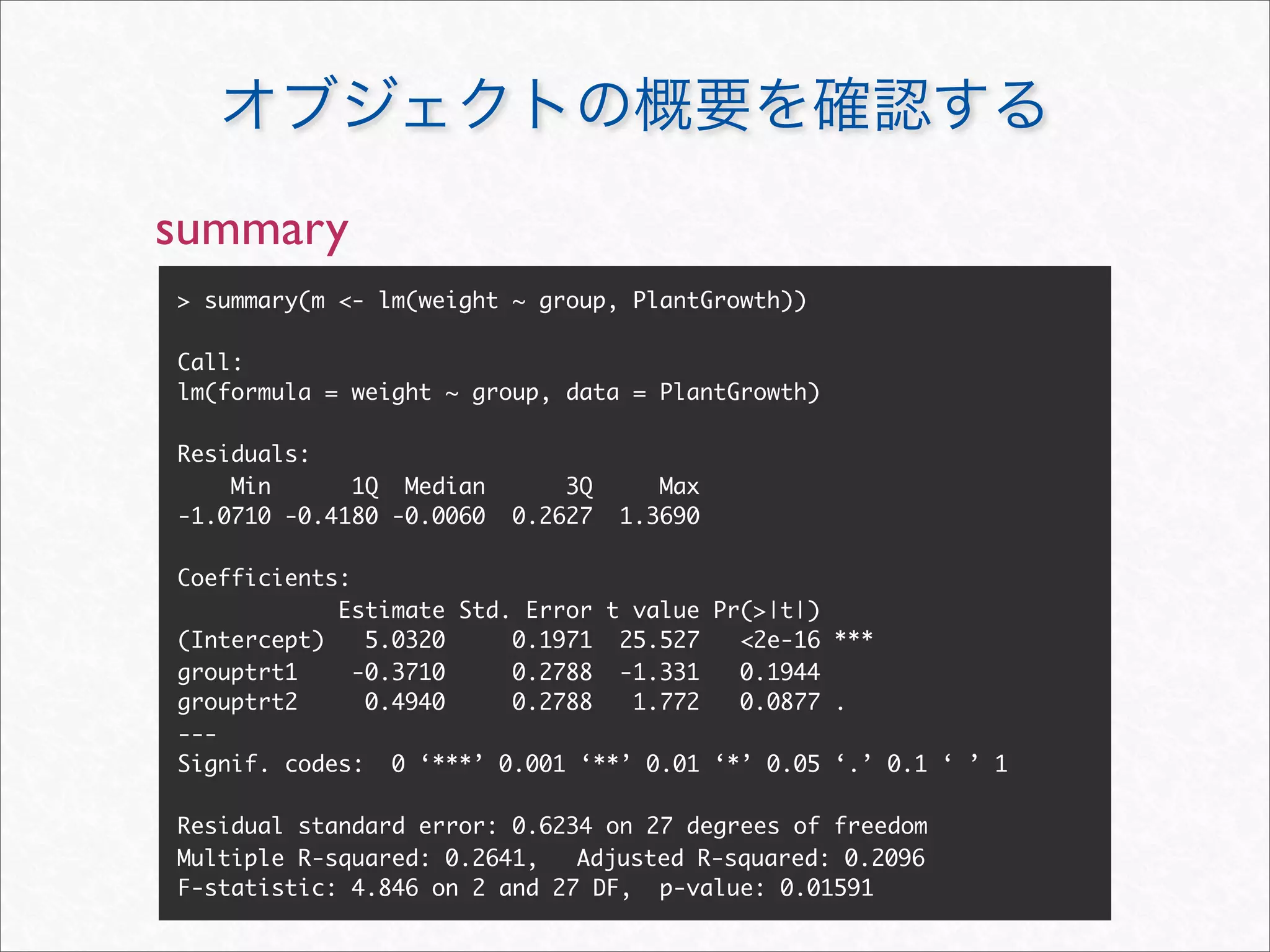
![str
> m # print.lm(m)
Call:
lm(formula = weight ~ group, data = PlantGrowth)
Coefficients:
(Intercept) grouptrt1 grouptrt2
5.032 -0.371 0.494
> str(m)
List of 13
$ coefficients : Named num [1:3] 5.032 -0.371 0.494
..- attr(*, "names")= chr [1:3] "(Intercept)" "grouptrt1"
"grouptrt2"
> m$coefficients #
(Intercept) grouptrt1 grouptrt2
5.032 -0.371 0.494](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-25-2048.jpg)
![class
> class(m)
[1] "lm"
data.frame matrix
character factor
> (x <- c(b = "this is b", a = "this is a"))
b a
"this is b" "this is a"
> (index <- factor(c("a", "b")))
[1] a b
Levels: a b
> x[index] #
b a
"this is b" "this is a"
> x[as.character(index)] #
a b
"this is a" "this is b"](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-26-2048.jpg)



![selectMethod, methods
> selectMethod("coef", signature = "lm") # signature
Method Definition (Class "derivedDefaultMethod"):
function (object, ...)
UseMethod("coef")
<environment: namespace:stats>
Signatures:
object
target "ANY"
defined "ANY"
> methods(coef) # UseMethod methods
[1] coef.aov* coef.Arima* coef.default* coef.listof*
coef.nls*
Non-visible functions are asterisked
> coef.default # coef.lm coef.default
Error: object 'coef.default' not found](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-30-2048.jpg)
![selectMethod, methods
> selectMethod("coef", signature = "lm") # signature
Method Definition (Class "derivedDefaultMethod"):
function (object, ...)
UseMethod("coef")
<environment: namespace:stats>
Signatures:
object
target "ANY"
defined "ANY"
> methods(coef) # UseMethod methods
[1] coef.aov* coef.Arima* coef.default* coef.listof*
coef.nls*
*
Non-visible functions are asterisked
> coef.default # coef.lm coef.default
Error: object 'coef.default' not found](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-31-2048.jpg)
![package:::func
> stats:::coef.default # Non-visible pkg:::func
function (object, ...)
object$coefficients
<environment: namespace:stats>
> evalq(coef.default, environment(lm)) #
function (object, ...)
object$coefficients
<environment: namespace:stats>
> debug(stats:::coef.default)
> coef(m)
debugging in: coef.default(m)
debug: object$coefficients
Browse[2]>](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-32-2048.jpg)



![trace
trace 3 5
f <- function(x) { > trace(f, browser, at = c(3, 5))
cat("1st linen") > f()
if (TRUE) { 1st line
cat("1st 2nd linen") Tracing f() step 3
cat("2nd 2nd linen") Called from: eval(expr, envir,
} enclos)
Browse[1]> c
cat("3rd line 1st 2nd line
") 2nd 2nd line
cat("4th linen") 3rd line
} Tracing f() step 5
Called from: eval(expr, envir,
enclos)
Browse[1]> c
4th line](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-36-2048.jpg)
![trace
trace 3 5
f <- function(x) { > trace(f, browser, at = c(3, 5))
cat("1st linen") > f()
if (TRUE) { 1st line
cat("1st 2nd linen") Tracing f() step 3
cat("2nd 2nd linen") Called from: eval(expr, envir,
} enclos)
Browse[1]> c
cat("3rd line 1st 2nd line
") 2nd 2nd line
cat("4th linen") 3rd line
} Tracing f() step 5
Called from: eval(expr, envir,
enclos)
Browse[1]> c
4th line](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-37-2048.jpg)
![trace
trace f
f <- function(x) { # 1 > untrace(f) # trace
cat("1st linen") # 2 > as.list(body(f))
if (TRUE) { # 3 [[1]]
cat("1st 2nd linen") `{`
cat("2nd 2nd linen")
} # 1 1 [[2]]
# cat("1st linen")
cat("3rd line
[[3]]
") # 1 # 4
if (TRUE) {
cat("4th linen") # 5 cat("1st 2nd linen")
}
cat("2nd 2nd linen")
}
[[4]]
cat("3rd linen")
[[5]]
cat("4th linen")](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-38-2048.jpg)
![trace
trace trace
f <- function(x) { > trace(f, browser, at = c(3, 5))
cat("1st linen") [1] "f"
if (TRUE) { > body(f)
cat("1st 2nd linen") {
cat("2nd 2nd linen") cat("1st linen")
} {
.doTrace(browser(), "step 3")
cat("3rd line if (TRUE) {
") cat("1st 2nd linen")
cat("4th linen") cat("2nd 2nd linen")
} }
}
cat("3rd linen")
{
.doTrace(browser(), "step 5")
cat("4th linen")
}
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-39-2048.jpg)
![trace
trace trace
f <- function(x) { > trace(f, browser, at = c(3, 5))
cat("1st linen") [1] "f"
if (TRUE) { > body(f)
cat("1st 2nd linen") {
cat("2nd 2nd linen") cat("1st linen")
} { .doTrace
.doTrace(browser(), "step 3")
cat("3rd line if (TRUE) {
") cat("1st 2nd linen")
cat("4th linen") cat("2nd 2nd linen")
} }
}
cat("3rd linen")
{
.doTrace(browser(), "step 5")
cat("4th linen")
}
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-40-2048.jpg)
![trace
trace trace
f <- function(x) { > f()
cat("1st linen") 1st line
if (TRUE) { Tracing f() step 3
cat("1st 2nd linen") Called from: eval(expr, envir,
cat("2nd 2nd linen") enclos)
} Browse[1]> Q
> # .doTrace
cat("3rd line > tracingState(FALSE)
") [1] TRUE
cat("4th linen") > f()
} 1st line
1st 2nd line
2nd 2nd line
3rd line
4th line
> # .doTrace
> tracingState(TRUE)
[1] FALSE](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-41-2048.jpg)
![trace
trace
f <- function(x) { > trace(f, edit = TRUE) # f
cat("1st linen") Waiting for Emacs...
if (TRUE) { [1] "f"
cat("1st 2nd linen") > body(f)
cat("2nd 2nd linen") {
} cat("1st linen")
cat("3rd linen")
cat("3rd line browser()
") cat("4th linen")
cat("4th linen") }
} > f()
1st line
3rd line
Called from: f()
f <- function(x) { Browse[1]>
cat("1st linen")
cat("3rd linen") ※ .doTrace
browser()
cat("4th linen") tracingState
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-42-2048.jpg)
![recover
recover
f <- function(x) { > f(1)
x <- g(x)
return(x) Enter a frame number, or 0 to exit
}
1: f(1)
g <- function(x) { 2: g(x)
y <- x + 1 3: h(y)
y <- h(y)
return(y) Selection: 1 # f(1)
} Called from: top level
Browse[1]> x
h <- function(y) { [1] 1
z <- y + 1
recover()
return(z)
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-43-2048.jpg)
![recover
recover
f <- function(x) { Browse[1]> c
x <- g(x)
return(x) Enter a frame number, or 0 to exit
}
1: f(1)
g <- function(x) { 2: g(x)
y <- x + 1 3: h(y)
y <- h(y)
return(y) Selection: 2 # g(x)
} Called from: f(1)
Browse[1]> y
h <- function(y) { [1] 2
z <- y + 1
recover()
return(z)
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-44-2048.jpg)
![recover
recover
f <- function(x) { Browse[1]> c
x <- g(x)
return(x) Enter a frame number, or 0 to exit
}
1: f(1)
g <- function(x) { 2: g(x)
y <- x + 1 3: h(y)
y <- h(y)
return(y) Selection: 3 # h(y)
} Called from: g(x)
Browse[1]> z
h <- function(y) { [1] 3
z <- y + 1
recover()
return(z)
}](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-45-2048.jpg)
![options(error = recover)
recover recover
f <- function(x) { > options(error = recover)
x <- g(x) > f(1)
return(x) Error in print(x) : object 'x' not
} found
g <- function(x) { Enter a frame number, or 0 to exit
y <- x + 1
y <- h(y) 1: f(1)
return(y) 2: g(x)
} 3: h(y)
4: print(x)
h <- function(y) {
z <- y + 1 Selection: 3 # h(y)
print(x) # Error! Called from: h(y)
return(z) Browse[1]> x
} Error during wrapup: object 'x'
not found
Browse[1]> ls()
[1] "y" "z"](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-46-2048.jpg)
![options(error = dump.frames)
dump.frames
dump.frames recover
f <- function(x) { > options(error = dump.frames)
x <- g(x) > f(1)
return(x) Error in print(x) : object 'x' not
} found
> ls()
g <- function(x) { [1] "f" "g" "h" "last.dump"
y <- x + 1 > debugger(last.dump)
y <- h(y) Message: Error in print(x) :
return(y) object 'x' not found
} Available environments had calls:
1: f(1)
h <- function(y) { 2: g(x)
z <- y + 1 3: h(y)
print(x) # Error! 4: print(x)
return(z)
} Enter an environment number, or 0
to exit Selection:](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-47-2048.jpg)

![ksvm
> library(kernlab)
> ksvm(Species ~ .,
+ iris,
+ class.weights = c(versicolor = 1, setosa = 10),
+ prob.model = TRUE)
Using automatic sigma estimation (sigest) for RBF or laplace
kernel
Error in lev(ret)[yd[cind]] :
only 0's may be mixed with negative subscripts](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-49-2048.jpg)
![ksvm
> options(error = recover) # recover
> ksvm(Species ~ .,
+ iris,
+ class.weights = c(versicolor = 1, setosa = 10),
+ prob.model = TRUE)
Using automatic sigma estimation (sigest) for RBF or laplace kernel
Error in lev(ret)[yd[cind]] :
only 0's may be mixed with negative subscripts
Enter a frame number, or 0 to exit
1: ksvm(Species ~ ., iris, class.weights = c(versicolor = 1, setosa
= 10), pro
< >
9: .local(x, ...)
10: as.factor(lev(ret)[yd[cind]])
11: is.factor(x)
12: inherits(x, "factor")
Selection: 10](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-50-2048.jpg)
![ksvm
Called from: top level
Browse[1]> ls()
[1] "x"
Browse[1]> x
Error during wrapup: promise already under evaluation: recursive
default argument reference or earlier problems?](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-51-2048.jpg)
![ksvm
Called from: top level
Browse[1]> ls()
[1] "x"
Browse[1]> x
Error during wrapup: promise already under evaluation: recursive
default argument reference or earlier problems?](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-52-2048.jpg)
![ksvm
> options(error = NULL)
> ksvm(Species ~ .,
+ iris,
+ class.weights = c(versicolor = 1, setosa = 10),
+ prob.model = TRUE)
Using automatic sigma estimation (sigest) for RBF or laplace kernel
Error in lev(ret)[yd[cind]] :
only 0's may be mixed with negative subscripts
> ksvm #
standardGeneric for "ksvm" defined from package "kernlab"
function (x, ...)
standardGeneric("ksvm")
<environment: 0x103623e08>
Methods may be defined for arguments: x
Use showMethods("ksvm") for currently available ones.
S4](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-53-2048.jpg)

![ksvm
> selectMethod(ksvm, "matrix")
Method Definition:
function (x, ...)
{
.local <- function (x, y = NULL, scaled = TRUE, type = NULL,
kernel = "rbfdot", kpar = "automatic", C = 1, nu = 0.2,
< >
, drop = FALSE][cind, ], as.factor(lev(ret)
[yd[cind]]),
< >](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-55-2048.jpg)
![ksvm
> trace(ksvm, signature = “matrix”, edit = TRUE)
browser
browser()
if (is.null(class.weights))
cret <- ksvm(x[c(indexes[[i]], indexes
[[j]]),
, drop = FALSE][cind, ], yd[cind],
type = type(ret), kernel = kernel,
kpar = NULL, C = C, nu = nu, tol = tol,
scaled = FALSE, cross = 0, fit = FALSE,
cache = cache, prob.model = FALSE)
else cret <- ksvm(x[c(indexes[[i]], indexes
[[j]]),
, drop = FALSE][cind, ], as.factor(lev(ret)
[yd[cind]]),](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-56-2048.jpg)
![ksvm
> ksvm(Species ~ .,
+ iris,
+ class.weights = c(versicolor = 1, setosa = 10),
+ prob.model = TRUE)
Using automatic sigma estimation (sigest) for RBF or laplace kernel
Called from: .local(x, ...)
Browse[1]> yd
[1] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[22] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[43] -1 -1 -1 -1 -1 -1 -1 -1 1 1 1 1 1 1 1 1 1 1 1 1 1
[64] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[85] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
lev(ret)[yd[cind]] yd (-1, 1)
yd lev(ret)](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-57-2048.jpg)

![> 1:5
[1] 1 2 3 4 5
> .Last.value
[1] 1 2 3 4 5
ESS TAB .Last.value](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-59-2048.jpg)
![f <- function(x) {
y <- x
g(y)
}
g <- function(x) {
x <- x^2
browser()
}
> f(10)
Called from: g(y) # x y
Browse[1]> x # x 2
[1] 100
Browse[1]> y # y
Error: object 'y' not found](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-60-2048.jpg)
![Browse[1]> evalq(y, parent.frame()) # y
[1] 10
Browse[1]> evalq(ls(), parent.frame()) #
[1] "x" "y"
Browse[1]> eval.parent(quote(y)) # part1
[1] 10
Browse[1]> get("y", parent.frame()) # part2
[1] 10
Browse[1]> ls(envir = parent.frame()) #
[1] "x" "y"
evalq(expr, parent.frame())](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-61-2048.jpg)

![apply browser
> debug(f)
> f()
debugging in: f()
debug: {
a <- lapply(1:10000, function(x) {
browser()
print(x)
})
}
Browse[2]> n
debug: a <- lapply(1:10000, function(x) {
browser()
print(x)
})
Browse[2]>
Called from: FUN(1:10000[[1L]], ...)
Browse[2]> c
[1] 1
Called from: FUN(1:10000[[2L]], ...) # browser()
Browse[2]>](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-63-2048.jpg)
![apply browser
Browse[2]> evalq(FUN <- edit(FUN), parent.frame())
function(x) {
browser() #
print(x)
}
Browse[2]> evalq(fix(FUN), parent.frame())
Browse[2]> FUN <<- evalq(edit(FUN), parent.frame())](https://image.slidesharecdn.com/rdebug-110827082208-phpapp01/75/R-64-2048.jpg)