Transcription .pdf
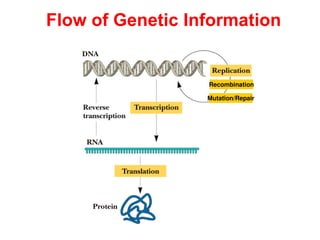
- 1. Flow of Genetic Information Recombination Mutation/Repair
- 2. Three Different Classes of RNA 1) rRNA (ribosomal) • large (long) RNA molecules • structural and functional components of ribosomes • highly abundant 2) mRNA (messenger) • typically small (short) • encode proteins • multiple types, not abundant 3) tRNA (transfer) and small ribosomal RNAs • very small • Important in translation Not all genes encode proteins
- 3. In Bacteria - all three classes are transcribed by the same RNA polymerase In Eukaryotes - each class is transcribed by a different RNA Polymerase •RNAP I - rRNAs •RNAP II - mRNAs •RNAP III - tRNAs & small ribosomal RNAs Different Types of RNA Polymerase
- 4. (coding strand) (sense strand) (non-coding strand) (anti-sense strand) RNAP
- 5. 5’ 5’ 3’ 3’ Template strand +1 Transcription Initiation Site Direction of transcription “Downstream” “Upstream” +2 +3 +4 +5 +6 -3 -1 -2 -4 -5 There is no “zero”
- 6. •Promoters - DNA sequences that guide RNAP to the beginning of a gene (transcription initiation site). •Terminators - DNA sequences that specify then termination of RNA synthesis and release of RNAP from the DNA. •RNA Polymerase (RNAP) - Enzyme for synthesis of RNA. •Steps • 1) Initiation. 2) Elongation. 3) Termination. Bacterial (Prokaryotic) Transcription
- 7. -10 region RNAP binds a region of DNA from -40 to +20 The sequence of the non-template strand is shown TTGACA…16-19 bp... TATAAT “-35” spacer “-10”
- 8. Important Promoter Features • The closer the match to the consensus the stronger the promoter (-10 and -35 boxes) • The absolute sequence of the spacer region (between the -10 and -35 boxes) is not important • The length of the spacer sequence IS important: TTGACA - spacer (16 to 19 base pairs) - TATAAT • Spacers that are longer or shorter than the consensus length make weak promoters
- 9. Properties of Promoters • Promoters typically consist of a 40 bp region on the 5'-side of the transcription start site • Two consensus sequence elements: – The "-35 region", with consensus TTGACA – The Pribnow box near -10, with consensus TATAAT - this region is ideal for unwinding.
- 10. RNA Polymerase Has Many Functions • Scan DNA and identify promoters • Bind to promoters • Initiate transcription • Elongate the RNA chain • Terminate transcription • Be responsive to regulatory proteins (activators and repressors) Thus, RNAP is a multisubunit enzyme
- 11. Transcription in Prokaryotes • In E.coli, RNA polymerase is a 465 kD complex, with 2 , 1 , 1 ', 1 (holoenzyme). • Core enzyme is 2 , 1 , 1 ’ (can transcribe but it can’t find promoters). • recognizes promoter sequences on DNA; ' binds DNA; binds NTPs and interacts with . subunits appear to be essential for assembly and for activation of enzyme by regulatory proteins.
- 12. 2 2 2’ = core enzyme I ’ II CORE ENZYME Sequence-independent, nonspecific transcription initiation + vegetative (principal ) 70 SIGMA SUBUNIT interchangeable, promoter recognition The assembly pathway of the core enzyme heat shock (for emergencies) 32 nitrogen starvation (for emergencies) 60
- 13. I ’ II 70 RNAP HOLOENZYME -70 Promoter-specific transcription initiation In the Holoenzyme: · ' binds DNA · binds NTPs · and ' together make up the active site · subunits appear to be essential for assembly and for activation of enzyme by regulatory proteins. They also bind DNA. · recognizes promoter sequences on DNA
- 14. Binding of polymerase to Template DNA • Polymerase binds nonspecifically to DNA with low affinity and migrates, looking for promoter. • Sigma subunit recognizes promoter sequence. • RNA polymerase holoenzyme and promoter form "closed promoter complex" (DNA not unwound). • Polymerase unwinds about 12 base pairs (A-T rich) to form "open promoter complex“.
- 15. Finding and binding the promoter Closed complex formation RNAP bound -40 to +20 Open complex formation RNAP unwinds from - 10 to +2 Binding of 1st NTP Requires high purine [NTP] Addition of next NTPs Requires lower purine [NTPs] Dissociation of sigma After RNA chain is 6-10 NTPs long
- 16. Chain Elongation Core polymerase - no sigma • Polymerase is accurate - only about 1 error in 10,000 bases (not as accurate as DNAP III) • Even this error rate negligible- since many transcripts are made from each gene • Elongation rate is 20-50 bases per second - slower in G/C-rich regions and faster elsewhere • Topoisomerases precede and follow polymerase to relieve supercoiling
- 17. Two mechanisms 1. Rho dependent Rho - the termination factor protein An ATP-dependent helicase – it moves along the RNA transcript, finds the "bubble", unwinds it and releases the RNA chain. Chain Termination
- 18. Rho-Dependent Transcription Termination (depends on a protein AND a DNA sequence) G/C -rich site RNAP slows down Rho helicase catches up Elongating complex is disrupted
- 19. The termination function of factor The factor, a hexamer, is a ATPase and a helicase.
- 20. Two mechanisms 2) Rho-Independent (Intrinsic) - termination sites in DNA – inverted repeat, rich in G:C, which forms a stem- loop in RNA transcript – 6-8 nos of A’s in DNA coding for U’s in transcript Chain Termination
- 21. Rho-Independent Transcription Termination (depends on DNA sequence - NOT a protein factor) Stem-loop structure
- 22. Rho-independent transcription termination • RNAP pauses when it reaches a termination site. • The pause may give the hairpin structure time to fold • The fold disrupts important interactions between the RNAP and its RNA product • The U-rich RNA can dissociate from the template • The complex is now disrupted and elongation is terminated
- 23. • Bacterial environment changes rapidly. • Survival depends on ability to adapt. • Bacteria must express the enzymes required to survive in that environment. • Enzyme synthesis is costly (energetically). • So make enzymes only when required. Transcription Regulation in Prokaryotes Why is it necessary?
- 24. Proteins Constitutive • Always expressed • “housekeeping” • e.g. glucose metabolizing enzymes • Glucose is the preferred carbon source for bacteria Adaptive • “inducible” • Made only when needed • e.g. Lactose metabolizing enzymes • Made only if lactose is the sole carbon source • Not made if glucose is present
- 25. 1. Alternate sigma factor usage: controls selective transcription of entire sets of genes s32 s60 vegetative (principal s) heat shock nitrogen starvation s70 TTGACA TATAAT (16-19 bp) (5-9 bp) A +1 CNCTTGA CCCATNT (13-15 bp) (5-9 bp) A +1 CTGGNA TTGCA (6 bp) (5-9 bp) A +1 Ways to Regulate Transcription
- 26. 2. Positive Regulation (activation): a positive regulatory factor (activator) improves the ability of RNAP to bind to and initiate transcription at a weak promoter. Ways to Regulate Transcription RNAP -35 -10 +1 Activator Activator binding site EXAMPLE: CAP
- 27. 3. Negative Regulation (repression): a negative regulatory factor (repressor) blocks the ability of RNAP to bind to and initiate transcription at a strong promoter. Ways to Regulate Transcription RNAP -35 -10 Repressor Operator EXAMPLE: lac REPRESSOR
- 28. Protein Synthesis is Regulated Transcriptionally • Genes that encode proteins with related functions are grouped into transcriptional units called “operons” • This ensures that genes for enzymes in the same metabolic pathway are all made at the same time Operons have three functional “parts” 1) structural genes: these encode proteins 2) promoter 3) regulatory sequences that interact with regulatory proteins Sometimes an operon is associated with: 4) regulatory genes: these encode proteins regulating expression of that operon
- 29. Structural genes promoter Operator (regulatory sequence that binds a repressor protein) Architecture of a typical operon By regulating a single promoter you can co-ordinate the expression of three genes (in this example) RNA transcript covers all genes in the operon = “polycistronic RNA”
- 31. RNA polymerases – Much more complex than prokaryotic RNAP a) RNAP I – synthesizes ribosomal RNA b) RNAP II – synthesizes messenger RNA c) RNAP III – synthesizes transfer RNA and 1 type of rRNA Eukaryotic RNAPs have subunits that are homologous to a, b, and b’ of prokaryotic RNAP; however, eukaryotic RNAP also contain many additional subunits.
- 32. a. Initiation • Transcription initiation needs promoter and upstream regulatory regions. • The cis-acting elements are the specific sequences on the DNA template that regulate the transcription of one or more genes.
- 33. Eukaryotic promoters • a) contain a TATA rich region located –25 to -30 from the start of transcription • b) CCAAT (frequently at –75) • c) GC box • d) Some promoters have other sequences located either upstream or downstream that maximize the level of transcription called enhancers
- 34. structural gene GCGC CAAT TATA intron exon exon start CAAT box GC box enhancer cis-acting element TATA box (Hogness box) Cis-acting element
- 35. TATA box
- 36. • RNA-pol does not bind the promoter directly. • RNA-pol II associates with six transcription factors, TFII A - TFII H. • The trans-acting factors are the proteins that recognize and bind directly or indirectly cis-acting elements and regulate its activity. Transcription factors
- 37. TF for eukaryotic transcription
- 38. Assembly of RNA pol and transcription factors at promoter • Formation of closed complex –TATA binding protein (TBP) binds to TATA box. • TBP requires TFIIB • TFIIA can stabilize TFIIB-TBP complex. • To TFIIB-TBP complex binds another complex , TFIIF and Pol II. • TFIIF helps target Pol II to promoters.. • TFIIE and TFIIH bind to form the closed complex. • TFIIH has helicase activity- unwinds DNA
- 39. Pre-initiation complex (PIC) RNA pol II TF II F TBP TAF TATA DNA TF II A TF II B TF II E TF II H
- 40. • TF II H is of protein kinase activity to phosphorylate CTD of RNA-pol. • (CTD is the C-terminal domain of RNA-pol) • Only the p-RNA-pol can move toward the downstream, starting the elongation phase. • Most of the TFs fall off from PIC during the elongation phase. Phosphorylation of RNA-pol
- 41. • The elongation is similar to that of prokaryotes. • The transcription and translation do not take place simultaneously since they are separated by nuclear membrane. b. Elongation
- 42. • The termination sequence is AATAAA followed by GT repeats. • The termination is closely related to the post-transcriptional modification. c. Termination